
Rice Science ›› 2019, Vol. 26 ›› Issue (6): 393-403.DOI: 10.1016/j.rsci.2018.11.002
• Research Paper • Previous Articles Next Articles
Pathaichindachote Wanwarang1,2,3, Panyawut Natjaree4, Sikaewtung Kannika4, Patarapuwadol Sujin1,2,5, Muangprom Amorntip1,2,4( )
)
Received:2018-07-10
Accepted:2018-11-06
Online:2019-11-28
Published:2019-08-19
Pathaichindachote Wanwarang, Panyawut Natjaree, Sikaewtung Kannika, Patarapuwadol Sujin, Muangprom Amorntip. Genetic Diversity and Allelic Frequency of Selected Thai and Exotic Rice Germplasm Using SSR Markers[J]. Rice Science, 2019, 26(6): 393-403.
Add to citation manager EndNote|Ris|BibTeX
| Noa | Rice accession | Agro-ecologies and special traitb | Noa | Rice accessions | Agro-ecologies and special traitsb | Noa | Rice accessions | Agro-ecologies and special traitsb |
|---|---|---|---|---|---|---|---|---|
| 1 | Abhayae | Resistance to Brown planthopper | 57 | Niaw Pang Pahd | Upland, Colored rice | 113 | Bueng Bawng Wod | Upland rice, Central |
| 2 | Azucenag | Tropical japonica, Resistance to Drought | 58 | OIC 27 IIId | Upland, Colored rice | 114 | Bueng Choo Bang Pueyd | Upland rice, Central |
| 3 | Nipponbareg | Temperate japonica | 59 | Chaw Maid | Upland, Colored rice, South | 115 | Gra Thingd | Upland rice, Central |
| 4 | Pokkalig | Resistance to Salt/ Brown planthopper | 60 | Dam Raid | Upland, Colored rice, South | 116 | Huay Muangd | Upland rice, Central |
| 5 | Rathu Heenati e | Resistance to Brown planthopper | 61 | Graw Sawd | Upland, Colored rice, South | 117 | Lep Mood | Upland rice, Central |
| 6 | Taichung Native 1e | High-yielding, semi-dwarf rice | 62 | Hawm Nin-capsf | Lowland, Colored rice | 118 | Meed | Upland rice, Central |
| 7 | IR 13429-150-3-2-1-2g | Resistance to Gall midge | 63 | Hawm Nin-Plueak Khao Ton Khiaof | Lowland, Colored rice | 119 | Non Raid | Upland rice, Central |
| 8 | IR 1905-PP11-29-4-61g | Resistance to Blast | 64 | Hawm Nin-Plueak Muangf | Lowland, Colored rice | 120 | Pan Aung Jerngd | Upland rice, Central |
| 9 | IR 20g | New Plant type, Stem borer | 65 | Sang Yod Thung Ku Laf | Lowland, Colored rice | 121 | Pan Maw Wand | Upland rice, Central |
| 10 | IR 28g | Earliness, Multiple resistance | 66 | Sang Yod Phat Tha Lungc | Lowland, Colored rice, Photosensitive | 122 | Pra Plenrgd | Upland rice, Central |
| 11 | IR 4422-98-3-6-1g | Resistance to Salt | 67 | Gam Kaf | Colored rice | 123 | Toh Tia Uad | Upland rice, Central |
| 12 | IR 4630-22-2-17g | Resistance to Salt | 68 | Gam Pef | Colored rice | 124 | Ton Tia Guay Nud | Upland rice, Central |
| 13 | IR 48g | Multiple resistance | 69 | Gam Ton Khiawf | Colored rice | 125 | Gra Bawkd | Upland rice, North |
| 14 | IR 52 (IR 5853-118-5) g | Resistance to Brown planthopper | 70 | 4806d | Upland rice | 126 | Ja Ge Wahd | Upland rice, North |
| 15 | IR 5533 - PP854 - 1g | Resistance to Blast | 71 | Bahn Daengd | Upland rice | 127 | Jao Hawmd | Upland rice, North |
| 16 | IR 58g | Earliness | 72 | Baw Dend | Upland rice | 128 | Jao Khahwd | Upland rice, North |
| 17 | IR 6023-10-1-1g | New Plant type, Drought Resistance | 73 | Chao Nahng Prungd | Upland rice | 129 | Jao Li Saw San Pah Tongc | Upland rice, North |
| 18 | IR 64g | Long grain parboiled rice, good eating quality | 74 | Daw Dawk Phraod | Upland rice | 130 | Ngaw Pind | Upland rice, North |
| 19 | IR 68544-29-2-1-3-1-2e | Tropical Japonica, New Plant Type, High-yielding | 75 | Gahb Damd | Upland rice | 131 | Sew Mae Chanc | Upland rice, North |
| 20 | IR 72g | Earliness | 76 | Gam Piak Khahwf | Upland rice | 132 | Chao Kaod | Upland rice, North east |
| 21 | IR 8608-298-3-1g | Resistance to Bacterial blight | 77 | Jam Pah Tawng Noid | Upland rice | 133 | Dok Makd | Upland rice, North east |
| 22 | IR 9-60g | Resistance to Brown planthopper | 78 | Jam Pahd | Upland rice | 134 | E Daengd | Upland rice, North east |
| 23 | IR 9884-54-3g | Resistance to Salt, Multiple resistance | 79 | Jek Chueyd | Upland rice | 135 | E Phaed | Upland rice, North east |
| 24 | Bue Lomd | Upland, Colored rice | 80 | Kahp Yaof | Upland rice | 136 | Jao Tah Hin Ngomd | Upland rice, North east |
| 25 | Cawm Plid | Upland, Colored rice | 81 | Khao Daengd | Upland rice | 137 | Jao Tep Pah Ratd | Upland rice, North east |
| 26 | Daw Damd | Upland, Colored rice (dark purple stripe green leaves, dark purple stalk, brown speckle and black grain), and Fragrance | 82 | Khao Gonf | Upland rice | 138 | Tah Gua Pahd | Upland rice, North east |
| 27 | Daw Mueyd | Upland, Colored rice | 83 | Khao Gra Bid | Upland rice | 139 | Dam Hawmd | Upland rice, South |
| 28 | Dawk Dod | Upland, Colored rice | 84 | Khao Hawmd | Upland rice | 140 | Dam Lungd | Upland rice, South |
| 29 | Dawk La Mudd | Upland, Colored rice | 85 | Khao Khun Maed | Upland rice | 141 | Dawk Mudd | Upland rice, South |
| 30 | E God | Upland, Colored rice | 86 | Khao Klahd | Upland rice | 142 | Dawk Pah Yawmc | Upland rice, South, Resistance to Blast, Brown spot, Narrow brown spot, Fragrance, Good cooking quality |
| 31 | Gam Doi Sa Kedc | Upland, Colored rice | 87 | Khao Loid | Upland rice | 143 | Dawk Pud Raid | Upland rice, South |
| 32 | Gamd | Upland, Colored rice | 88 | Khao Lueang d | Upland rice | 144 | Goo Mueang Luangc | Upland rice, South, Resistance to Drought, Blast and Brown spot, Narrow brown spot |
| 33 | Gamd | Upland, Colored rice | 89 | Khao Ma Led Lekd | Upland rice | 145 | Jae Sand | Upland rice, South |
| 34 | Glamd | Upland, Colored rice | 90 | Khao Niawd | Upland rice | 146 | Jaekd | Upland rice, South |
| 35 | GP 33 CAC 155d | Upland, Colored rice | 91 | Khao Piangd | Upland rice | 147 | Jao Je Wahd | Upland rice, South |
| 36 | Haed | Upland, Colored rice | 92 | Khao Plah Laid | Upland rice | 148 | Jee Mah Leed | Upland rice, South |
| 37 | Ja Pah Gawd | Upland, Colored rice | 93 | Khao Puangd | Upland rice | 149 | Sai Raid | Upland rice, South |
| 38 | Khao Daengd | Upland, Colored rice | 94 | Khao Ruang Yaod | Upland, Fragrance | 150 | Sue Rahd | Upland rice, South |
| 39 | Khao Gamd | Upland, Colored rice | 95 | Khao Sa Ahdd | Upland rice | 151 | Tho Maid | Upland rice, South |
| 40 | Khao Niaw Damd | Upland, Colored rice | 96 | Khao Set Tid | Upland rice | 152 | Chiang Phat Tha Lungc | Lowland, Rainfed for South, Good milling quality |
| 41 | Khao Niaw Damd | Upland, Colored rice, Fragrance | 97 | Khao Tah Kluebd | Upland rice | 153 | Kai Noif | Lowland rice |
| 42 | Lai Mahkd | Upland, Colored rice | 98 | Khao Thua Damd | Upland rice | 154 | KDML105e | Lowland, Breeding line, Photosensitive rainfed lowland rice, good eating quality, desirable fragrance |
| 43 | Lang Gaid | Upland, Colored rice | 99 | Khiaw Nok Gra Lingd | Upland rice | 155 | Khai Mod Rin 3c | Lowland rice, Photosensitive |
| 44 | Lueang Noid | Upland, Colored rice | 100 | Khiaw Nok Gra Lingd | Upland, Fragrance | 156 | Khao Luang San Pah Tongc | Lowland rice, Photosensitive |
| 45 | Lueang Thawngd | Upland, Colored rice | 101 | Lueang Noid | Upland rice | 157 | Khao Ngah Changc | Lowland rice, Resistance to salt |
| 46 | Mae Look Awnd | Upland, Colored rice | 102 | Lueang Tawngd | Upland rice | 158 | Khao Pra Du Daengc | Lowland rice, Photosensitive |
| 47 | Nang Khiawd | Upland, Colored rice | 103 | Ma Li Awngd | Upland rice | 159 | Khem Thong Phat Tha Lungc | Lowland rice, Photosensitive |
| 48 | Niaw Dam Hawmd | Upland, Colored rice | 104 | Ma Li Leuayd | Upland rice | 160 | KLG96006-76-1c | Lowland rice, Breeding line |
| 49 | Niaw Dam Noid | Upland, Colored rice, Fragrance | 105 | Nahng Ngahmd | Upland rice | 161 | Leb Nok Pattanic | Lowland, Rainfed for South, Late flowering, Good cooking quality |
| 50 | Niaw Damd | Upland, Colored rice | 106 | Niaw Look Puengd | Upland rice | 162 | Lueang Thong-N21f | Lowland rice |
| 51 | Niaw Damd | Upland, Colored rice | 107 | Pong Tiad | Upland rice | 163 | Nahm Sa Gui 19c | Lowland, Rainfed for North and North East, Early flowering, Resistance to Brown planthopper |
| 52 | Niaw Damd | Upland, Colored rice | 108 | Puang Tawngd | Upland rice | 164 | PTT1c | Lowland, Improved line, Irrigated, Fragrance, High yielding |
| 53 | Niaw Di Tah Ked | Upland, Colored rice | 109 | Ruang Diawd | Upland rice | 165 | RD11c | Lowland, Improved line, Non-photosensitive, Resistance to Blast, Resistance to Brown spot |
| 54 | Niaw Ga Am Mah Kad | Upland, Colored rice | 110 | Tah Pibd | Upland rice | 166 | RD39c | Lowland, Improved line, Non-photosensitive, Resistance to Blast, good cooking quality |
| 55 | Niaw Gan Yahd | Upland, Colored rice | 111 | Tawng Mah Engd | Upland rice | 167 | SPR91062-5-PTT-1-2-1c | Lowland rice, Breeding line |
| 56 | Niaw Lan Tand | Upland, Colored rice | 112 | Bue Kha Nongd | Upland rice, Central |
Supplemental Table 1 Characteristics of rice accession used in the study with their agro-ecologies and special traits.
| Noa | Rice accession | Agro-ecologies and special traitb | Noa | Rice accessions | Agro-ecologies and special traitsb | Noa | Rice accessions | Agro-ecologies and special traitsb |
|---|---|---|---|---|---|---|---|---|
| 1 | Abhayae | Resistance to Brown planthopper | 57 | Niaw Pang Pahd | Upland, Colored rice | 113 | Bueng Bawng Wod | Upland rice, Central |
| 2 | Azucenag | Tropical japonica, Resistance to Drought | 58 | OIC 27 IIId | Upland, Colored rice | 114 | Bueng Choo Bang Pueyd | Upland rice, Central |
| 3 | Nipponbareg | Temperate japonica | 59 | Chaw Maid | Upland, Colored rice, South | 115 | Gra Thingd | Upland rice, Central |
| 4 | Pokkalig | Resistance to Salt/ Brown planthopper | 60 | Dam Raid | Upland, Colored rice, South | 116 | Huay Muangd | Upland rice, Central |
| 5 | Rathu Heenati e | Resistance to Brown planthopper | 61 | Graw Sawd | Upland, Colored rice, South | 117 | Lep Mood | Upland rice, Central |
| 6 | Taichung Native 1e | High-yielding, semi-dwarf rice | 62 | Hawm Nin-capsf | Lowland, Colored rice | 118 | Meed | Upland rice, Central |
| 7 | IR 13429-150-3-2-1-2g | Resistance to Gall midge | 63 | Hawm Nin-Plueak Khao Ton Khiaof | Lowland, Colored rice | 119 | Non Raid | Upland rice, Central |
| 8 | IR 1905-PP11-29-4-61g | Resistance to Blast | 64 | Hawm Nin-Plueak Muangf | Lowland, Colored rice | 120 | Pan Aung Jerngd | Upland rice, Central |
| 9 | IR 20g | New Plant type, Stem borer | 65 | Sang Yod Thung Ku Laf | Lowland, Colored rice | 121 | Pan Maw Wand | Upland rice, Central |
| 10 | IR 28g | Earliness, Multiple resistance | 66 | Sang Yod Phat Tha Lungc | Lowland, Colored rice, Photosensitive | 122 | Pra Plenrgd | Upland rice, Central |
| 11 | IR 4422-98-3-6-1g | Resistance to Salt | 67 | Gam Kaf | Colored rice | 123 | Toh Tia Uad | Upland rice, Central |
| 12 | IR 4630-22-2-17g | Resistance to Salt | 68 | Gam Pef | Colored rice | 124 | Ton Tia Guay Nud | Upland rice, Central |
| 13 | IR 48g | Multiple resistance | 69 | Gam Ton Khiawf | Colored rice | 125 | Gra Bawkd | Upland rice, North |
| 14 | IR 52 (IR 5853-118-5) g | Resistance to Brown planthopper | 70 | 4806d | Upland rice | 126 | Ja Ge Wahd | Upland rice, North |
| 15 | IR 5533 - PP854 - 1g | Resistance to Blast | 71 | Bahn Daengd | Upland rice | 127 | Jao Hawmd | Upland rice, North |
| 16 | IR 58g | Earliness | 72 | Baw Dend | Upland rice | 128 | Jao Khahwd | Upland rice, North |
| 17 | IR 6023-10-1-1g | New Plant type, Drought Resistance | 73 | Chao Nahng Prungd | Upland rice | 129 | Jao Li Saw San Pah Tongc | Upland rice, North |
| 18 | IR 64g | Long grain parboiled rice, good eating quality | 74 | Daw Dawk Phraod | Upland rice | 130 | Ngaw Pind | Upland rice, North |
| 19 | IR 68544-29-2-1-3-1-2e | Tropical Japonica, New Plant Type, High-yielding | 75 | Gahb Damd | Upland rice | 131 | Sew Mae Chanc | Upland rice, North |
| 20 | IR 72g | Earliness | 76 | Gam Piak Khahwf | Upland rice | 132 | Chao Kaod | Upland rice, North east |
| 21 | IR 8608-298-3-1g | Resistance to Bacterial blight | 77 | Jam Pah Tawng Noid | Upland rice | 133 | Dok Makd | Upland rice, North east |
| 22 | IR 9-60g | Resistance to Brown planthopper | 78 | Jam Pahd | Upland rice | 134 | E Daengd | Upland rice, North east |
| 23 | IR 9884-54-3g | Resistance to Salt, Multiple resistance | 79 | Jek Chueyd | Upland rice | 135 | E Phaed | Upland rice, North east |
| 24 | Bue Lomd | Upland, Colored rice | 80 | Kahp Yaof | Upland rice | 136 | Jao Tah Hin Ngomd | Upland rice, North east |
| 25 | Cawm Plid | Upland, Colored rice | 81 | Khao Daengd | Upland rice | 137 | Jao Tep Pah Ratd | Upland rice, North east |
| 26 | Daw Damd | Upland, Colored rice (dark purple stripe green leaves, dark purple stalk, brown speckle and black grain), and Fragrance | 82 | Khao Gonf | Upland rice | 138 | Tah Gua Pahd | Upland rice, North east |
| 27 | Daw Mueyd | Upland, Colored rice | 83 | Khao Gra Bid | Upland rice | 139 | Dam Hawmd | Upland rice, South |
| 28 | Dawk Dod | Upland, Colored rice | 84 | Khao Hawmd | Upland rice | 140 | Dam Lungd | Upland rice, South |
| 29 | Dawk La Mudd | Upland, Colored rice | 85 | Khao Khun Maed | Upland rice | 141 | Dawk Mudd | Upland rice, South |
| 30 | E God | Upland, Colored rice | 86 | Khao Klahd | Upland rice | 142 | Dawk Pah Yawmc | Upland rice, South, Resistance to Blast, Brown spot, Narrow brown spot, Fragrance, Good cooking quality |
| 31 | Gam Doi Sa Kedc | Upland, Colored rice | 87 | Khao Loid | Upland rice | 143 | Dawk Pud Raid | Upland rice, South |
| 32 | Gamd | Upland, Colored rice | 88 | Khao Lueang d | Upland rice | 144 | Goo Mueang Luangc | Upland rice, South, Resistance to Drought, Blast and Brown spot, Narrow brown spot |
| 33 | Gamd | Upland, Colored rice | 89 | Khao Ma Led Lekd | Upland rice | 145 | Jae Sand | Upland rice, South |
| 34 | Glamd | Upland, Colored rice | 90 | Khao Niawd | Upland rice | 146 | Jaekd | Upland rice, South |
| 35 | GP 33 CAC 155d | Upland, Colored rice | 91 | Khao Piangd | Upland rice | 147 | Jao Je Wahd | Upland rice, South |
| 36 | Haed | Upland, Colored rice | 92 | Khao Plah Laid | Upland rice | 148 | Jee Mah Leed | Upland rice, South |
| 37 | Ja Pah Gawd | Upland, Colored rice | 93 | Khao Puangd | Upland rice | 149 | Sai Raid | Upland rice, South |
| 38 | Khao Daengd | Upland, Colored rice | 94 | Khao Ruang Yaod | Upland, Fragrance | 150 | Sue Rahd | Upland rice, South |
| 39 | Khao Gamd | Upland, Colored rice | 95 | Khao Sa Ahdd | Upland rice | 151 | Tho Maid | Upland rice, South |
| 40 | Khao Niaw Damd | Upland, Colored rice | 96 | Khao Set Tid | Upland rice | 152 | Chiang Phat Tha Lungc | Lowland, Rainfed for South, Good milling quality |
| 41 | Khao Niaw Damd | Upland, Colored rice, Fragrance | 97 | Khao Tah Kluebd | Upland rice | 153 | Kai Noif | Lowland rice |
| 42 | Lai Mahkd | Upland, Colored rice | 98 | Khao Thua Damd | Upland rice | 154 | KDML105e | Lowland, Breeding line, Photosensitive rainfed lowland rice, good eating quality, desirable fragrance |
| 43 | Lang Gaid | Upland, Colored rice | 99 | Khiaw Nok Gra Lingd | Upland rice | 155 | Khai Mod Rin 3c | Lowland rice, Photosensitive |
| 44 | Lueang Noid | Upland, Colored rice | 100 | Khiaw Nok Gra Lingd | Upland, Fragrance | 156 | Khao Luang San Pah Tongc | Lowland rice, Photosensitive |
| 45 | Lueang Thawngd | Upland, Colored rice | 101 | Lueang Noid | Upland rice | 157 | Khao Ngah Changc | Lowland rice, Resistance to salt |
| 46 | Mae Look Awnd | Upland, Colored rice | 102 | Lueang Tawngd | Upland rice | 158 | Khao Pra Du Daengc | Lowland rice, Photosensitive |
| 47 | Nang Khiawd | Upland, Colored rice | 103 | Ma Li Awngd | Upland rice | 159 | Khem Thong Phat Tha Lungc | Lowland rice, Photosensitive |
| 48 | Niaw Dam Hawmd | Upland, Colored rice | 104 | Ma Li Leuayd | Upland rice | 160 | KLG96006-76-1c | Lowland rice, Breeding line |
| 49 | Niaw Dam Noid | Upland, Colored rice, Fragrance | 105 | Nahng Ngahmd | Upland rice | 161 | Leb Nok Pattanic | Lowland, Rainfed for South, Late flowering, Good cooking quality |
| 50 | Niaw Damd | Upland, Colored rice | 106 | Niaw Look Puengd | Upland rice | 162 | Lueang Thong-N21f | Lowland rice |
| 51 | Niaw Damd | Upland, Colored rice | 107 | Pong Tiad | Upland rice | 163 | Nahm Sa Gui 19c | Lowland, Rainfed for North and North East, Early flowering, Resistance to Brown planthopper |
| 52 | Niaw Damd | Upland, Colored rice | 108 | Puang Tawngd | Upland rice | 164 | PTT1c | Lowland, Improved line, Irrigated, Fragrance, High yielding |
| 53 | Niaw Di Tah Ked | Upland, Colored rice | 109 | Ruang Diawd | Upland rice | 165 | RD11c | Lowland, Improved line, Non-photosensitive, Resistance to Blast, Resistance to Brown spot |
| 54 | Niaw Ga Am Mah Kad | Upland, Colored rice | 110 | Tah Pibd | Upland rice | 166 | RD39c | Lowland, Improved line, Non-photosensitive, Resistance to Blast, good cooking quality |
| 55 | Niaw Gan Yahd | Upland, Colored rice | 111 | Tawng Mah Engd | Upland rice | 167 | SPR91062-5-PTT-1-2-1c | Lowland rice, Breeding line |
| 56 | Niaw Lan Tand | Upland, Colored rice | 112 | Bue Kha Nongd | Upland rice, Central |
| Chr. | Marker | Sequence of primer | Product size | Repeat motif |
|---|---|---|---|---|
| 1 | RM283 | F: GTCTACATGTACCCTTGTTGGG R: CGGCATGAGAGTCTGTGATG | 151 | (GA)18 |
| 2 | RM5529 | F: AGCCGAAACTACATTCGGTG R: TTGTGTAGTTGGCACGCTTC | 167 | (TG)13 |
| 3 | RM7 | F: TTCGCCATGAAGTCTCTCG R: CCTCCCATCATTTCGTTGTT | 180 | (GA)19 |
| 3 | RM313 | F: TGCTACAAGTGTTCTTCAGGAC R: GCTCACCTTTTGTGTTCCAC | 111 | (GT)6CA(CG)5-6-(GT)8 |
| 4 | RM317 | F: CATACTTACCAGTTCACCGCC R: CTGGAGAGTGTCAGCTAGTTGA | 155 | (GC)4(GT)18 |
| 5 | RM169 | F: TGGCTGGCTCCGTGGGTAGCTG R: TCCCGTTGCCGTTCATCCCTCC | 167 | (GA)12 |
| 6 | RM588 | F: GTTGCTCTGCCTCACTCTTG R: AACGAGCCAACGAAGCAG | 126 | (TGC)9 |
| 7 | RM542 | F: TGAATCAAGCCCCTCACTAC R: CTGCAACGAGTAAGGCAGAG | 113 | (CT)22 |
| 8 | RM1381 | F: AATGCTTGCATTCGTTACAC R: TTAAATGGGAGTTCTTGTGC | 236 | (AG)33 |
| 9 | RM108 | F: TCTCTTGCGCGCACACTGGCAC R: CGTGCACCACCACCACCACCAC | 80 | (GGT)10 |
| 10 | RM496 | F: GACATGCGAACAACGACATC R: GCTGCGGCGCTGTTATAC | 267 | (TC)14 |
| 11 | RM21 | F: ACAGTATTCCGTAGGCACGG R: GCTCCATGAGGGTGGTAGAG | 157 | (GA)18 |
| 12 | RM247 | F: TAGTGCCGATCGATGTAACG R: CATATGGTTTTGACAAAGCG | 131 | (CT)16 |
Supplemental Table 2 Thirteen SSR markers with their sequences selected for the study.
| Chr. | Marker | Sequence of primer | Product size | Repeat motif |
|---|---|---|---|---|
| 1 | RM283 | F: GTCTACATGTACCCTTGTTGGG R: CGGCATGAGAGTCTGTGATG | 151 | (GA)18 |
| 2 | RM5529 | F: AGCCGAAACTACATTCGGTG R: TTGTGTAGTTGGCACGCTTC | 167 | (TG)13 |
| 3 | RM7 | F: TTCGCCATGAAGTCTCTCG R: CCTCCCATCATTTCGTTGTT | 180 | (GA)19 |
| 3 | RM313 | F: TGCTACAAGTGTTCTTCAGGAC R: GCTCACCTTTTGTGTTCCAC | 111 | (GT)6CA(CG)5-6-(GT)8 |
| 4 | RM317 | F: CATACTTACCAGTTCACCGCC R: CTGGAGAGTGTCAGCTAGTTGA | 155 | (GC)4(GT)18 |
| 5 | RM169 | F: TGGCTGGCTCCGTGGGTAGCTG R: TCCCGTTGCCGTTCATCCCTCC | 167 | (GA)12 |
| 6 | RM588 | F: GTTGCTCTGCCTCACTCTTG R: AACGAGCCAACGAAGCAG | 126 | (TGC)9 |
| 7 | RM542 | F: TGAATCAAGCCCCTCACTAC R: CTGCAACGAGTAAGGCAGAG | 113 | (CT)22 |
| 8 | RM1381 | F: AATGCTTGCATTCGTTACAC R: TTAAATGGGAGTTCTTGTGC | 236 | (AG)33 |
| 9 | RM108 | F: TCTCTTGCGCGCACACTGGCAC R: CGTGCACCACCACCACCACCAC | 80 | (GGT)10 |
| 10 | RM496 | F: GACATGCGAACAACGACATC R: GCTGCGGCGCTGTTATAC | 267 | (TC)14 |
| 11 | RM21 | F: ACAGTATTCCGTAGGCACGG R: GCTCCATGAGGGTGGTAGAG | 157 | (GA)18 |
| 12 | RM247 | F: TAGTGCCGATCGATGTAACG R: CATATGGTTTTGACAAAGCG | 131 | (CT)16 |
| Marker | Chromosome | Ng | Na | He | Ho | PIC |
|---|---|---|---|---|---|---|
| RM283 | 1 | 7 | 5 | 0.39 | 0.02 | 0.35 |
| RM5529 | 2 | 7 | 6 | 0.62 | 0.01 | 0.58 |
| RM7 | 3 | 6 | 6 | 0.64 | 0.00 | 0.60 |
| RM313 | 3 | 5 | 4 | 0.43 | 0.01 | 0.38 |
| RM317 | 4 | 9 | 9 | 0.69 | 0.00 | 0.64 |
| RM169 | 5 | 11 | 11 | 0.80 | 0.00 | 0.77 |
| RM588 | 6 | 5 | 4 | 0.39 | 0.07 | 0.33 |
| RM542 | 7 | 9 | 8 | 0.27 | 0.01 | 0.26 |
| RM1381 | 8 | 29 | 19 | 0.83 | 0.07 | 0.82 |
| RM108 | 9 | 4 | 4 | 0.58 | 0.00 | 0.51 |
| RM496 | 10 | 8 | 7 | 0.44 | 0.01 | 0.40 |
| RM21 | 11 | 19 | 16 | 0.87 | 0.02 | 0.86 |
| RM247 | 12 | 13 | 11 | 0.77 | 0.01 | 0.75 |
| Mean | 10.2 | 8.5 | 0.59 | 0.02 | 0.56 | |
| Total | 132 | 110 | ||||
Table 1 Genetic parameters of 13 simple sequence repeat markers in 167 rice accessions.
| Marker | Chromosome | Ng | Na | He | Ho | PIC |
|---|---|---|---|---|---|---|
| RM283 | 1 | 7 | 5 | 0.39 | 0.02 | 0.35 |
| RM5529 | 2 | 7 | 6 | 0.62 | 0.01 | 0.58 |
| RM7 | 3 | 6 | 6 | 0.64 | 0.00 | 0.60 |
| RM313 | 3 | 5 | 4 | 0.43 | 0.01 | 0.38 |
| RM317 | 4 | 9 | 9 | 0.69 | 0.00 | 0.64 |
| RM169 | 5 | 11 | 11 | 0.80 | 0.00 | 0.77 |
| RM588 | 6 | 5 | 4 | 0.39 | 0.07 | 0.33 |
| RM542 | 7 | 9 | 8 | 0.27 | 0.01 | 0.26 |
| RM1381 | 8 | 29 | 19 | 0.83 | 0.07 | 0.82 |
| RM108 | 9 | 4 | 4 | 0.58 | 0.00 | 0.51 |
| RM496 | 10 | 8 | 7 | 0.44 | 0.01 | 0.40 |
| RM21 | 11 | 19 | 16 | 0.87 | 0.02 | 0.86 |
| RM247 | 12 | 13 | 11 | 0.77 | 0.01 | 0.75 |
| Mean | 10.2 | 8.5 | 0.59 | 0.02 | 0.56 | |
| Total | 132 | 110 | ||||
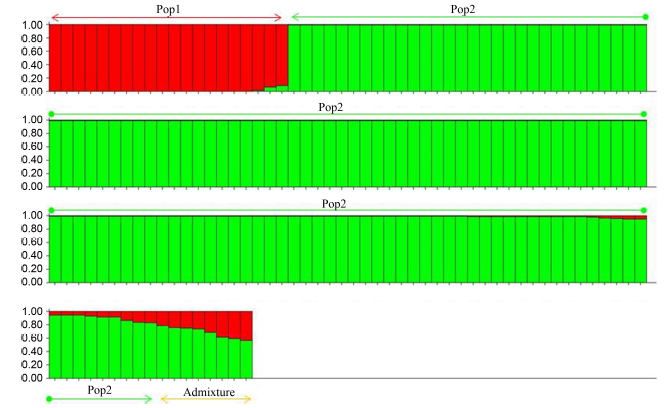
Fig. 4. Admixture model-based population structure based on K as 2.Each thin bar represents one accession. Red color code represents japonica proportion of the individuals and green color code represents indica proportion of the individuals. The length of color code in each bar indicates the proportion of the estimated membership coefficient.
| No. | Rice accession | Fi | No. | Rice accession | Fi | No. | Rice accession | Fi |
|---|---|---|---|---|---|---|---|---|
| 1 | Abhaya | 0.58 | 57 | Niaw Pang Pah | 0.31 | 113 | Bueng Bawng Wo | 0.04 |
| 2 | Azucena | 0.04 | 58 | OIC 27 III | 0.54 | 114 | Bueng Choo Bang Puey | 0.19 |
| 3 | Nipponbare | 0.19 | 59 | Chaw Mai | 0.42 | 115 | Gra Thing | 0.58 |
| 4 | Pokkali | 0.19 | 60 | Dam Rai | 0.38 | 116 | Huay Muang | 0.19 |
| 5 | Rathu Heenati | 0.35 | 61 | Graw Saw | 0.46 | 117 | Lep Moo | 0.77 |
| 6 | Taichung Native 1 | 0.38 | 62 | Hawm Nin-caps | 0.50 | 118 | Mee | 0.54 |
| 7 | IR13429-150-3-2-1-2 | 0.27 | 63 | Hawm Nin-Plueak Khao Ton Khiao | 0.65 | 119 | Non Rai | 1.00 |
| 8 | IR1905-PP11-29-4-61 | 0.54 | 64 | Hawm Nin-Plueak Muang | 0.50 | 120 | Pan Aung Jerng | 0.42 |
| 9 | IR20 | 0.50 | 65 | Sang Yod Thung Ku La | 0.54 | 121 | Pan Maw Wan | 0.04 |
| 10 | IR28 | 0.58 | 66 | Sang Yod Phat Tha Lung | 0.54 | 122 | Pra Plenrg | 0.50 |
| 11 | IR4422-98-3-6-1 | 0.35 | 67 | Gam Ka | 0.50 | 123 | Toh Tia Ua | 0.12 |
| 12 | IR4630-22-2-17 | 0.42 | 68 | Gam Pe | 0.50 | 124 | Ton Tia Guay Nu | 0.04 |
| 13 | IR48 | 0.50 | 69 | Gam Ton Khiaw | 0.65 | 125 | Gra Bawk | 0.08 |
| 14 | IR52 (IR5853-118-5) | 0.38 | 70 | 4806 | 0.04 | 126 | Ja Ge Wah | 0.50 |
| 15 | IR5533-PP854-1 | 0.50 | 71 | Bahn Daeng | 0.46 | 127 | Jao Hawm | 0.58 |
| 16 | IR58 | 0.50 | 72 | Baw Den | 0.35 | 128 | Jao Khahw | 0.19 |
| 17 | IR6023-10-1-1 | 0.42 | 73 | Chao Nahng Prung | 0.54 | 129 | Jao Li Saw San Pah Tong | 0.19 |
| 18 | IR64 | 0.62 | 74 | Daw Dawk Phrao | 0.58 | 130 | Ngaw Pin | 0.35 |
| 19 | IR68544-29-2-1-3-1-2 | 0.12 | 75 | Gahb Dam | 0.46 | 131 | Sew Mae Chan | 0.50 |
| 20 | IR72 | 0.58 | 76 | Gam Piak Khahw | 0.54 | 132 | Chao Kao | 0.58 |
| 21 | IR8608-298-3-1 | 0.58 | 77 | Jam Pah Tawng Noi | 0.58 | 133 | Dok Mak | 0.23 |
| 22 | IR9-60 | 0.42 | 78 | Jam Pah | 0.54 | 134 | E Daeng | 0.50 |
| 23 | IR9884-54-3 | 0.35 | 79 | Jek Chuey | 0.46 | 135 | E Phae | 0.50 |
| 24 | Bue Lom | 0.46 | 80 | Kahp Yao | 0.50 | 136 | Jao Tah Hin Ngom | 0.12 |
| 25 | Cawm Pli | 0.46 | 81 | Khao Daeng-No.81 | 0.46 | 137 | Jao Tep Pah Rat | 0.54 |
| 26 | Daw Dam | 0.50 | 82 | Khao Gon | 0.62 | 138 | Tah Gua Pah | 0.35 |
| 27 | Daw Muey | 0.62 | 83 | Khao Gra Bi | 0.54 | 139 | Dam Hawm | 0.46 |
| 28 | Dawk Do | 0.38 | 84 | Khao Hawm | 0.65 | 140 | Dam Lung | 0.12 |
| 29 | Dawk La Mud | 0.50 | 85 | Khao Khun Mae | 0.50 | 141 | Dawk Mud | 0.62 |
| 30 | E Go | 0.46 | 86 | Khao Klah | 0.46 | 142 | Dawk Pah Yawm | 0.19 |
| 31 | Gam Doi Sa Ked | 0.54 | 87 | Khao Loi | 0.50 | 143 | Dawk Pud Rai | 0.19 |
| 32 | Gam | 0.35 | 88 | Khao Lueang | 0.46 | 144 | Goo Mueang Luang | 0.73 |
| 33 | Gam | 0.54 | 89 | Khao Ma Led Lek | 0.46 | 145 | Jae San | 0.46 |
| 34 | Glam | 0.62 | 90 | Khao Niaw | 0.58 | 146 | Jaek | 0.50 |
| 35 | GP 33 CAC 155 | 0.42 | 91 | Khao Piang | 0.50 | 147 | Jao Je Wah | 0.19 |
| 36 | Hae | 0.42 | 92 | Khao Plah Lai | 0.62 | 148 | Jee Mah Lee | 0.31 |
| 37 | Ja Pah Gaw | 0.50 | 93 | Khao Puang | 0.35 | 149 | Sai Rai | 0.19 |
| 38 | Khao Daeng | 0.46 | 94 | Khao Ruang Yao | 0.69 | 150 | Sue Rah | 0.42 |
| 39 | Khao Gam | 0.58 | 95 | Khao Sa Ahd | 0.42 | 151 | Tho Mai | 0.38 |
| 40 | Khao Niaw Dam | 0.65 | 96 | Khao Set Ti | 0.58 | 152 | Chiang Phat Tha Lung | 0.35 |
| 41 | Khao Niaw Dam | 0.54 | 97 | Khao Tah Klueb | 0.46 | 153 | Kai Noi | 0.46 |
| 42 | Lai Mahk | 0.38 | 98 | Khao Thua Dam | 0.50 | 154 | KDML105 | 0.65 |
| 43 | Lang Gai | 0.58 | 99 | Khiaw Nok Gra Ling | 0.62 | 155 | Khai Mod Rin 3 | 0.42 |
| 44 | Lueang Noi | 0.50 | 100 | Khiaw Nok Gra Ling | 0.65 | 156 | Khao Luang San Pah Tong | 0.35 |
| 45 | Lueang Thawng | 0.42 | 101 | Lueang Noi | 0.54 | 157 | Khao Ngah Chang | 0.35 |
| 46 | Mae Look Awn | 0.38 | 102 | Lueang Tawng | 0.42 | 158 | Khao Pra Du Daeng | 0.58 |
| 47 | Nang Khiaw | 0.62 | 103 | Ma Li Awng | 0.46 | 159 | Khem Thong Phat Tha Lung | 0.54 |
| 48 | Niaw Dam Hawm | 0.12 | 104 | Ma Li Leuay | 0.58 | 160 | KLG96006-76-1 | 0.62 |
| 49 | Niaw Dam Noi | 0.69 | 105 | Nahng Ngahm | 0.54 | 161 | Leb Nok Pattani | 0.27 |
| 50 | Niaw Dam | 0.50 | 106 | Niaw Look Pueng | 0.50 | 162 | Lueang Thong-N21 | 0.50 |
| 51 | Niaw Dam | 0.42 | 107 | Pong Tia | 0.35 | 163 | Nahm Sa Gui 19 | 0.54 |
| 52 | Niaw Dam | 0.62 | 108 | Puang Tawng | 0.58 | 164 | PTT1 | 0.58 |
| 53 | Niaw Di Tah Ke | 0.50 | 109 | Ruang Diaw | 0.62 | 165 | RD11 | 0.27 |
| 54 | Niaw Ga Am Mah Ka | 0.46 | 110 | Tah Pib | 0.65 | 166 | RD39 | 0.50 |
| 55 | Niaw Gan Yah | 0.46 | 111 | Tawng Mah Eng | 0.54 | 167 | SPR91062-5-PTT-1-2-1 | 0.62 |
| 56 | Niaw Lan Tan | 0.54 | 112 | Bue Kha Nong | 0.50 |
Supplemental Table 3 Frequency of indica allele (Fi) calculated based on fingerprint profiles of 13 SSR markers.
| No. | Rice accession | Fi | No. | Rice accession | Fi | No. | Rice accession | Fi |
|---|---|---|---|---|---|---|---|---|
| 1 | Abhaya | 0.58 | 57 | Niaw Pang Pah | 0.31 | 113 | Bueng Bawng Wo | 0.04 |
| 2 | Azucena | 0.04 | 58 | OIC 27 III | 0.54 | 114 | Bueng Choo Bang Puey | 0.19 |
| 3 | Nipponbare | 0.19 | 59 | Chaw Mai | 0.42 | 115 | Gra Thing | 0.58 |
| 4 | Pokkali | 0.19 | 60 | Dam Rai | 0.38 | 116 | Huay Muang | 0.19 |
| 5 | Rathu Heenati | 0.35 | 61 | Graw Saw | 0.46 | 117 | Lep Moo | 0.77 |
| 6 | Taichung Native 1 | 0.38 | 62 | Hawm Nin-caps | 0.50 | 118 | Mee | 0.54 |
| 7 | IR13429-150-3-2-1-2 | 0.27 | 63 | Hawm Nin-Plueak Khao Ton Khiao | 0.65 | 119 | Non Rai | 1.00 |
| 8 | IR1905-PP11-29-4-61 | 0.54 | 64 | Hawm Nin-Plueak Muang | 0.50 | 120 | Pan Aung Jerng | 0.42 |
| 9 | IR20 | 0.50 | 65 | Sang Yod Thung Ku La | 0.54 | 121 | Pan Maw Wan | 0.04 |
| 10 | IR28 | 0.58 | 66 | Sang Yod Phat Tha Lung | 0.54 | 122 | Pra Plenrg | 0.50 |
| 11 | IR4422-98-3-6-1 | 0.35 | 67 | Gam Ka | 0.50 | 123 | Toh Tia Ua | 0.12 |
| 12 | IR4630-22-2-17 | 0.42 | 68 | Gam Pe | 0.50 | 124 | Ton Tia Guay Nu | 0.04 |
| 13 | IR48 | 0.50 | 69 | Gam Ton Khiaw | 0.65 | 125 | Gra Bawk | 0.08 |
| 14 | IR52 (IR5853-118-5) | 0.38 | 70 | 4806 | 0.04 | 126 | Ja Ge Wah | 0.50 |
| 15 | IR5533-PP854-1 | 0.50 | 71 | Bahn Daeng | 0.46 | 127 | Jao Hawm | 0.58 |
| 16 | IR58 | 0.50 | 72 | Baw Den | 0.35 | 128 | Jao Khahw | 0.19 |
| 17 | IR6023-10-1-1 | 0.42 | 73 | Chao Nahng Prung | 0.54 | 129 | Jao Li Saw San Pah Tong | 0.19 |
| 18 | IR64 | 0.62 | 74 | Daw Dawk Phrao | 0.58 | 130 | Ngaw Pin | 0.35 |
| 19 | IR68544-29-2-1-3-1-2 | 0.12 | 75 | Gahb Dam | 0.46 | 131 | Sew Mae Chan | 0.50 |
| 20 | IR72 | 0.58 | 76 | Gam Piak Khahw | 0.54 | 132 | Chao Kao | 0.58 |
| 21 | IR8608-298-3-1 | 0.58 | 77 | Jam Pah Tawng Noi | 0.58 | 133 | Dok Mak | 0.23 |
| 22 | IR9-60 | 0.42 | 78 | Jam Pah | 0.54 | 134 | E Daeng | 0.50 |
| 23 | IR9884-54-3 | 0.35 | 79 | Jek Chuey | 0.46 | 135 | E Phae | 0.50 |
| 24 | Bue Lom | 0.46 | 80 | Kahp Yao | 0.50 | 136 | Jao Tah Hin Ngom | 0.12 |
| 25 | Cawm Pli | 0.46 | 81 | Khao Daeng-No.81 | 0.46 | 137 | Jao Tep Pah Rat | 0.54 |
| 26 | Daw Dam | 0.50 | 82 | Khao Gon | 0.62 | 138 | Tah Gua Pah | 0.35 |
| 27 | Daw Muey | 0.62 | 83 | Khao Gra Bi | 0.54 | 139 | Dam Hawm | 0.46 |
| 28 | Dawk Do | 0.38 | 84 | Khao Hawm | 0.65 | 140 | Dam Lung | 0.12 |
| 29 | Dawk La Mud | 0.50 | 85 | Khao Khun Mae | 0.50 | 141 | Dawk Mud | 0.62 |
| 30 | E Go | 0.46 | 86 | Khao Klah | 0.46 | 142 | Dawk Pah Yawm | 0.19 |
| 31 | Gam Doi Sa Ked | 0.54 | 87 | Khao Loi | 0.50 | 143 | Dawk Pud Rai | 0.19 |
| 32 | Gam | 0.35 | 88 | Khao Lueang | 0.46 | 144 | Goo Mueang Luang | 0.73 |
| 33 | Gam | 0.54 | 89 | Khao Ma Led Lek | 0.46 | 145 | Jae San | 0.46 |
| 34 | Glam | 0.62 | 90 | Khao Niaw | 0.58 | 146 | Jaek | 0.50 |
| 35 | GP 33 CAC 155 | 0.42 | 91 | Khao Piang | 0.50 | 147 | Jao Je Wah | 0.19 |
| 36 | Hae | 0.42 | 92 | Khao Plah Lai | 0.62 | 148 | Jee Mah Lee | 0.31 |
| 37 | Ja Pah Gaw | 0.50 | 93 | Khao Puang | 0.35 | 149 | Sai Rai | 0.19 |
| 38 | Khao Daeng | 0.46 | 94 | Khao Ruang Yao | 0.69 | 150 | Sue Rah | 0.42 |
| 39 | Khao Gam | 0.58 | 95 | Khao Sa Ahd | 0.42 | 151 | Tho Mai | 0.38 |
| 40 | Khao Niaw Dam | 0.65 | 96 | Khao Set Ti | 0.58 | 152 | Chiang Phat Tha Lung | 0.35 |
| 41 | Khao Niaw Dam | 0.54 | 97 | Khao Tah Klueb | 0.46 | 153 | Kai Noi | 0.46 |
| 42 | Lai Mahk | 0.38 | 98 | Khao Thua Dam | 0.50 | 154 | KDML105 | 0.65 |
| 43 | Lang Gai | 0.58 | 99 | Khiaw Nok Gra Ling | 0.62 | 155 | Khai Mod Rin 3 | 0.42 |
| 44 | Lueang Noi | 0.50 | 100 | Khiaw Nok Gra Ling | 0.65 | 156 | Khao Luang San Pah Tong | 0.35 |
| 45 | Lueang Thawng | 0.42 | 101 | Lueang Noi | 0.54 | 157 | Khao Ngah Chang | 0.35 |
| 46 | Mae Look Awn | 0.38 | 102 | Lueang Tawng | 0.42 | 158 | Khao Pra Du Daeng | 0.58 |
| 47 | Nang Khiaw | 0.62 | 103 | Ma Li Awng | 0.46 | 159 | Khem Thong Phat Tha Lung | 0.54 |
| 48 | Niaw Dam Hawm | 0.12 | 104 | Ma Li Leuay | 0.58 | 160 | KLG96006-76-1 | 0.62 |
| 49 | Niaw Dam Noi | 0.69 | 105 | Nahng Ngahm | 0.54 | 161 | Leb Nok Pattani | 0.27 |
| 50 | Niaw Dam | 0.50 | 106 | Niaw Look Pueng | 0.50 | 162 | Lueang Thong-N21 | 0.50 |
| 51 | Niaw Dam | 0.42 | 107 | Pong Tia | 0.35 | 163 | Nahm Sa Gui 19 | 0.54 |
| 52 | Niaw Dam | 0.62 | 108 | Puang Tawng | 0.58 | 164 | PTT1 | 0.58 |
| 53 | Niaw Di Tah Ke | 0.50 | 109 | Ruang Diaw | 0.62 | 165 | RD11 | 0.27 |
| 54 | Niaw Ga Am Mah Ka | 0.46 | 110 | Tah Pib | 0.65 | 166 | RD39 | 0.50 |
| 55 | Niaw Gan Yah | 0.46 | 111 | Tawng Mah Eng | 0.54 | 167 | SPR91062-5-PTT-1-2-1 | 0.62 |
| 56 | Niaw Lan Tan | 0.54 | 112 | Bue Kha Nong | 0.50 |
| [1] | Babu B K, Meena V, Agarwal V, Agrawal P K.2014. Population structure and genetic diversity analysis of Indian and exotic rice (Oryza sativa L.) accessions using SSR markers. Mol Biol Rep, 41(7): 4329-4339. |
| [2] | Bassam B J, Caetano-Anolles G, Gresshoff P M.1991. Fast and sensitive silver staining of DNA in polyacrylamide gels.Anal Biochem, 196(1): 80-83. |
| [3] | Bhattacharya K R, Ali S Z.2015. An Introduction to Rice-Grain Technology. New Delhi, India: Woodhead Publishing India Pvt. Ltd. |
| [4] | Botstein D, White R L, Skolnick M, Davis R W.1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms.Am J Hum Genet, 32(3): 314-331. |
| [5] | Chakhonkaen S, Pitnjam K, Saisuk W, Ukoskit K, Muangprom A.2012. Genetic structure of Thai rice and rice accessions obtained from the International Rice Research Institute.Rice, 5: 19-31. |
| [6] | Chang T T.1976. The origin, evolution, cultivation, dissemination, and diversification of Asian and African rices.Euphytica, 25(1): 425-441. |
| [7] | Chen C, Chen H, Shan J X, Zhu M Z, Shi M, Gao J P, Lin H X.2013. Genetic and physiological analysis of a novel type of interspecific hybrid weakness in rice.Mol Plant, 6(3): 716-728. |
| [8] | Dan Z W, Liu P, Huang W C, Zhou W, Yao G X, Hu J, Zhu R S, Lu B R, Zhu Y G.2014. Balance between a higher degree of heterosis and increased reproductive isolation: A strategic design for breeding inter-subspecific hybrid rice.PLoS One, 9(3): e93122. |
| [9] | Earl D A, vonHoldt B M.2012. Structure Harvester: A website and program for visualizing STRUCTURE output and implementing the Evanno method.Conserv Genet Resour, 4(2): 359-361. |
| [10] | Evanno G, Regnaut S, Goudet J.2005. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study.Mol Ecol, 14(8): 2611-2620. |
| [11] | Garris A J, Tai T H, Coburn J, Kresovich S, McCouch S.2005. Genetic structure and diversity in Oryza sativa L. Genetics, 169(3): 1631-1638. |
| [12] | Giarrocco L E, Marassi M A, Salerno G L.2007. Assessment of the genetic diversity in argentine rice cultivars with SSR markers.Crop Sci, 47(2): 853-858. |
| [13] | Glaszmann J C.1987. Isozymes and classification of Asian rice varieties.Theor Appl Genet, 74(1): 21-30. |
| [14] | Global Rice Science Partnership.2013. Rice Almanac. Los Baños, the Philippines: International Rice Research Institute. |
| [15] | Guo X, Elston R C.1999. Linkage information content of polymorphic genetic markers.Hum Hered, 49(2): 112-118. |
| [16] | Heal G, Walker B, Levin S, Arrow K, Dasgupta P, Daily G, Ehrlich P, Maler K G, Kautsky N, Lubchenco J, Schneider S, Starrett D.2004. Genetic diversity and interdependent crop choices in agriculture.Resour Energy Econ, 26(2): 175-184. |
| [17] | Hori K, Yamamoto T, Yano M.2017. Genetic dissection of agronomically important traits in closely related temperate japonica rice cultivars. Breeding Sci, 67(5): 427-434. |
| [18] | Hoshino A A, Bravo J P, Nobile P M, Morelli K A.2012. Microsatellites as tools for genetic diversity analysis, genetic diversity in microorganisms. InTechOpen: 382. |
| [19] | Jin L, Lu Y, Xiao P, Sun M, Corke H, Bao J S.2010. Genetic diversity and population structure of a diverse set of rice germplasm for association mapping.Theor Appl Genet, 121(3): 475-487. |
| [20] | Jones M P, Dingkuhn M, Alukosnm G K, Semon M.2004. InterspecificOryza sativa L. X O. Glaberrima Steud. progenies in upland rice improvement. Euphytica, 94(2): 237-246. |
| [21] | Kanawapee N, Sanitchon J, Srihaban P, Theerakulpisut P.2011. Genetic diversity analysis of rice cultivars (Oryza sativa L.) differing in salinity tolerance based on RAPD and SSR markers. Electron J Biotechnol, 14(6): 1-17. |
| [22] | Khush G S.1995. Breaking the yield frontier of rice.Geo Journal, 35(3): 329-332. |
| [23] | Khush G S.1997. Origin, dispersal, cultivation and variation of rice.Plant Mol Biol, 35: 25-34. |
| [24] | Khush G S.2001. Green revolution: The way forward.Nat Rev Genet, 2(10): 815-822. |
| [25] | Linares O F.2002. African rice (Oryza glaberrima): History and future potential. Proc Natl Acad Sci USA, 99: 16360-16365. |
| [26] | Liu K J, Muse S V.2005. PowerMarker: An integrated analysis environment for genetic marker analysis.Bioinformatics, 21(9): 2128-2129. |
| [27] | Liu P, Dan Z W, Wang Z, Li S Q, Li N W, Yan H X, Cai X X, Lu B R.2015. Predicting hybrid fertility from maker-based genetic divergence index of parental varieties: Implications for utilizing inter-subspecies heterosis in hybrid rice breeding.Euphytica, 203(1): 47-57. |
| [28] | Lu B R, Cai X X, Xin J.2009. Efficient indica and japonica rice identification based on the InDel molecular method: Its implication in rice breeding and evolutionary research. Prog Nat Sci, 19(10): 1241-1252. |
| [29] | Ma L, Zhu F G, Li Z W, Zhang J F, Li X, Dong J L, Wang T.2015. TALEN-based mutagenesis of lipoxygenase LOX3 enhances the storage tolerance of rice (Oryza sativa) seeds. PLoS One, 10(12): e0143877. |
| [30] | Mackill D J.1995. Classifyingjaponica rice cultivars with RAPD markers. Crop Sci, 35(3): 889-894. |
| [31] | Maclean J L.2002. Rice Almanac. England: CABI. |
| [32] | Mondini L, Noorani A, Pagnotta M A.2009. Assessing plant genetic diversity by molecular tools.Diversity, 1(1): 19-35. |
| [33] | Murray M G, Thompson W F.1980. Rapid isolation of high molecular weight plant DNA.Nucl Acids Res, 8(19): 4321-4325. |
| [34] | Nachimuthu V V, Muthurajan R, Duraialaguraja S, Sivakami R, Pandian B A, Ponniah G, Gunasekaran K, Swaminathan M, Suji K K, Sabariappan R.2015. Analysis of population structure and genetic diversity in rice germplasm using SSR markers: An initiative towards association mapping of agronomic traits inOryza sativa. Rice, 8(1): 30. |
| [35] | Nayar N M. 2014. Oryza species and their interrelationships. In: Origin and Phylogeny of Rices. San Diego: Academic Press: 59-115. |
| [36] | Nei M.1973. Analysis of gene diversity in subdivided populations.Proc Natl Acad Sci USA, 70(12): 3321-3323. |
| [37] | Ni J J, Colowit P M, Mackill D J.2002. Evaluation of genetic diversity in rice subspecies using microsatellite markers.Crop Sci, 42(2): 601-607. |
| [38] | Oka H I.1957. Genic analysis for the sterility of hybrids between distantly related varieties of cultivated rice.J Genet, 55(3): 397-409. |
| [39] | Oka H I.1958. Intervarietal variation and classification of cultivated rice.Ind J Genet Plant Breeding, 18: 79-89. |
| [40] | Oka H I.1988. Origin of Cultivated Rice. Amsterdam: Elsevier. |
| [41] | Panaud O, Chen X, McCouch S R.1996. Development of microsatellite markers and characterization of simple sequence length polymorphism (SSLP) in rice (Oryza sativa L.). Mol Gen Genet, 252(5): 597-607. |
| [42] | Peng S, Laza R C, Visperas R M, Khush G S, Virk P, Zhu D.2004. Rice: Progress in breaking the yield ceiling. New directions for a diverse planet. In: Proceedings of the 4th International Crop Science Congress. Australia: Brisbane. |
| [43] | Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A.1996. The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis.Mol Breeding, 2(3): 225-238. |
| [44] | Pritchard J K, Stephens M, Donnelly P.2000. Inference of population structure using multilocus genotype data.Genetics, 155(2): 945-959. |
| [45] | Ramanatha Rao V, Hodgkin T.2002. Genetic diversity and conservation and utilization of plant genetic resources.Plant Cell Tissue Organ Cult, 68(1): 1-19. |
| [46] | Rohlf F J.1998. NTSYSpc numerical taxonomy and multivariate analysis system version 2.0 user guide. Setauket, New York: Applied Biostatistics Inc. |
| [47] | Sakamoto T, Matsuoka M.2004. Generating high-yielding varieties by genetic manipulation of plant architecture.Curr Opin Biotechnol, 15(2): 144-147. |
| [48] | Salem K F M, Sallam A.2016. Analysis of population structure and genetic diversity of Egyptian and exotic rice (Oryza sativa L.) genotypes. CR Biol, 339(1): 1-9. |
| [49] | Singh N, Choudhury D R, Tiwari G, Singh A K, Kumar S, Srinivasan K, Tyagi R K, Sharma A D, Singh N K, Singh R.2016. Genetic diversity trend in Indian rice varieties: An analysis using SSR markers.BMC Genet, 17(1): 127. |
| [50] | Sneath P H A, Sokal R R.1973. Numerical Taxonomy. San Francisco, California, USA: W.H.: Freeman andCompany. |
| [51] | Subudhi P K, Sasaki T, Khush G S.2006. Rice, Cereals and Millets. Berlin, Heidelberg: Springer. |
| [52] | Sun C Q, Wang X K, Li Z C, Yoshimura A, Iwata N.2001. Comparison of the genetic diversity of common wild rice (Oryza rufipogon Griff.) and cultivated rice (O. sativa L.) using RFLP markers. Theor Appl Genet, 102(1): 157-162. |
| [53] | Sutoro, Lestari P, Kurniawan H.2015. Genetic diversity of upland rice landraces from java island as revealed by SSR markers.Indones J Agric Sci, 16(1): 1-10. |
| [54] | Tang S, Zhang Y, Zeng L, Luo L, Zhong Y, Geng Y.2010. Assessment of genetic diversity and relationships of upland rice accessions from southwest China using microsatellite markers.Plant Biosyst, 144(1): 85-92. |
| [55] | Thatsanabanjong K, Phoasavadi P.2017. The music of rice in Amphawa.J Silpakorn Univ: Veridian E, 10(5): 492-505. |
| [56] | Toro M A, Caballero A.2005. Characterization and conservation of genetic diversity in subdivided populations.Philos Trans R Soc Lond B Biol Sci, 360: 1367-1378. |
| [57] | Travis A J, Norton G J, Datta S, Sarma R, Dasgupta T, Savio F L, Macaulay M, Hedley P E, McNally K L, Sumon M H, Islam M R, Price A H.2015. Assessing the genetic diversity of rice originating from Bangladesh, Assam and West Bengal.Rice, 8(1): 35. |
| [58] | Virmani S S, Aquino R C, Khush G S.1982. Heterosis breeding in rice (Oryza sativa L.). Theor Appl Genet, 63(4): 373-380. |
| [59] | Wang C H, Zheng X M, Xu Q, Yuan X P, Huang L, Zhou H F, Wei X H, Ge S.2014. Genetic diversity and classification of Oryza sativa with emphasis on Chinese rice germplasm. Heredity, 112(5): 489-496. |
| [60] | Wang M M, Zhu Z F, Tan L B, Liu F X, Fu Y C, Sun C Q, Cai H W.2013. Complexity of indica-japonica varietal differentiation in Bangladesh rice landraces revealed by microsatellite markers. Breeding Sci, 63(2): 227-232. |
| [61] | Wang Y R, Qiu F L, Hua Z T, Dai G J.2010. Relationship of parental indica-japonica indexes with yield and grain quality traits of japonica hybrid rice in Northern China. Rice Sci, 17(3): 199-205. |
| [62] | Wuthiyano C.2000. Thai Rice Landraces. Pathum Thani, Thailand: Department of Agriculture. |
| [63] | Xu Q, Yuan X P, Wang S, Feng Y, Yu H Y, Wang Y P, Yang Y L, Wei X H, Li X M.2016. The genetic diversity and structure of indica rice in China as detected by single nucleotide polymorphism analysis. BMC Genet, 17: 53. |
| [64] | Yonemaru J I, Choi S H, Sakai H, Ando T, Shomura A, Yano M, Wu J, Fukuoka S.2015. Genome-wide indel markers shared by diverse Asian rice cultivars compared to Japanese rice cultivar ‘Koshihikari’.Breeding Sci, 65(3): 249-256. |
| [65] | Zhang D L, Zhang H L, Wei X H, Qi Y W, Wang M X, Sun J L, Ding L, Tang S X, Qiu Z E, Cao Y S, Wang X K, Li Z C.2007. Genetic structure and diversity of Oryza sativa L. in Guizhou, China. Chin Sci Bull, 52(3): 343-351. |
| [66] | Zhang L N, Cao G L, Han L Z.2013. Genetic diversity of rice landraces from lowland and upland accessions of China.Rice Sci, 20(4): 259-266. |
| [67] | Zhang P, Li J Q, Li X L, Liu X D, Zhao X J, Lu Y G.2011. Population structure and genetic diversity in a rice core collection (Oryza sativa L.) investigated with SSR markers. PLoS One, 6(12): e27565. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [13] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||