
Rice Science ›› 2019, Vol. 26 ›› Issue (6): 384-392.DOI: 10.1016/j.rsci.2018.09.002
• Research Paper • Previous Articles Next Articles
Yang Lv1,#, Yueying Wang1, Jahan Noushin1, Haitao Hu1, Ping Chen1, Lianguang Shang1, Haiyan Lin1, Guojun Dong1, Jiang Hu1, Zhenyu Gao1, Qian Qian1, Yu Zhang1,2( ), Longbiao Guo1(
), Longbiao Guo1( )
)
Received:2018-07-10
Accepted:2018-09-21
Online:2019-11-28
Published:2019-08-19
Contact:
Yang Lv
About author: These authors contributed equally to this work
Yang Lv, Yueying Wang, Jahan Noushin, Haitao Hu, Ping Chen, Lianguang Shang, Haiyan Lin, Guojun Dong, Jiang Hu, Zhenyu Gao, Qian Qian, Yu Zhang, Longbiao Guo. Genome-Wide Association Analysis and Allelic Mining of Grain Shape-Related Traits in Rice[J]. Rice Science, 2019, 26(6): 384-392.
Add to citation manager EndNote|Ris|BibTeX
| No. | Variety | Origin | No. | Variety | Origin | No. | Variety | Origin | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Hucchao1hao | Sichuan | 55 | Zhubao384 | Guangdong | 109 | Xianfeng1hao | Zhejiang | |||
| 2 | Hucheng17 | Sichuan | 56 | Baoxuan2hao | Guangxi | 110 | Zaolian31 | Zhejiang | |||
| 3 | Huhongzao1hao | Sichuan | 57 | Guangnan1hao | Guangxi | 111 | Zaoxian141 | Zhejiang | |||
| 4 | Hunanzao1hao | Sichuan | 58 | Hongnan | Guangxi | 112 | Zhechang3hao | Zhejiang | |||
| 5 | Hushuang1011 | Sichuan | 59 | Jiefang1hao | Guangxi | 113 | Zhengui9hao | Zhejiang | |||
| 6 | Aimakang | Sichuan | 60 | Jinguang202 | Guangxi | 114 | Zhenlong13 | Zhejiang | |||
| 7 | Aituogu151 | Sichuan | 61 | Tuanjie1hao | Guangxi | 115 | Zhenxian97B | Zhejiang | |||
| 8 | CDR22 | Sichuan | 62 | Nanjing1hao | Jiangsu | 116 | Zhongganzao | Zhejiang | |||
| 9 | Chengduai8hao | Sichuan | 63 | Qinglian16 | Jiangsu | 117 | Zhufei10hao | Zhejiang | |||
| 10 | Chenghui448 | Sichuan | 64 | Zhuxi26 | Jiangsu | 118 | Zhuke2hao | Zhejiang | |||
| 11 | Chuanxiang29B | Sichuan | 65 | 508 | Jiangxi | 119 | Zhulianai | Zhejiang | |||
| 12 | Fuhui838 | Sichuan | 66 | 5450 | Jiangxi | 120 | 1296 | Hubei | |||
| 13 | Gang46B | Sichuan | 67 | 6001 | Jiangxi | 121 | Ezao1hao | Hubei | |||
| 14 | K17B | Sichuan | 68 | 6004 | Jiangxi | 122 | Ezao6hao | Hubei | |||
| 15 | Shuhui527 | Sichuan | 69 | 6188 | Jiangxi | 123 | Ezhong2hao | Hubei | |||
| 16 | Yixiang1B | Sichuan | 70 | 7004 | Jiangxi | 124 | Huaai15 | Hubei | |||
| 17 | Annandao | Anhui | 71 | 7005 | Jiangxi | 125 | Jingainuo | Hubei | |||
| 18 | Dengdengqi | Anhui | 72 | Aihong11 | Jiangxi | 126 | Chun389B | Sichuan | |||
| 19 | Mafangxian | Anhui | 73 | Bayiwan | Jiangxi | 127 | Aizizhan | Hunan | |||
| 20 | Wuhu71zao | Anhui | 74 | Gannanzao20 | Jiangxi | 128 | Chenwan3hao | Hunan | |||
| 21 | XieqingzaoB | Anhui | 75 | Gannanzao23 | Jiangxi | 129 | Fanzi | Hunan | |||
| 22 | Zhuguang23 | Anhui | 76 | Jiangxisimiao | Jiangxi | 130 | Hongmidongzhan | Hunan | |||
| 23 | Aijiaonante | Guangdong | 77 | Jingnong3hao | Jiangxi | 131 | Hunanzao | Hunan | |||
| 24 | Conglu51 | Guangdong | 78 | Jingnong69 | Jiangxi | 132 | Laohuanggu | Hunan | |||
| 25 | Fengaizhan1 | Guangdong | 79 | Jiunongzao1hao | Jiangxi | 133 | Liguzao | Hunan | |||
| 26 | Guangchang13 | Guangdong | 80 | Liantangzao | Jiangxi | 134 | Luguiai | Hunan | |||
| 27 | Guangchangai | Guangdong | 81 | Nantehao | Jiangxi | 135 | Xiangaizao10hao | Hunan | |||
| 28 | Guangjie9hao | Guangdong | 82 | Wanxian22 | Jiangxi | 136 | Xiangaizao2hao | Hunan | |||
| 29 | Guangluai4hao | Guangdong | 83 | Xingheng1hao | Jiangxi | 137 | Xiangaizao4hao | Hunan | |||
| 30 | Guangnongai1hao | Guangdong | 84 | Xiujiangwan3hao | Jiangxi | 138 | Xiangaizao7hao | Hunan | |||
| 31 | Guangwen10hao | Guangdong | 85 | Xiujiangzao4hao | Jiangxi | 139 | Xiangaizao8hao | Hunan | |||
| 32 | Guangxuan3hao | Guangdong | 86 | Xiujiangzao9hao | Jiangxi | 140 | Xiangaizao9hao | Hunan | |||
| 33 | Guichao13 | Guangdong | 87 | Youzhanzi | Jiangxi | 141 | Xiangwanxian2hao | Hunan | |||
| 34 | Guichao2hao | Guangdong | 88 | 5006 | Henan | 142 | Xiangzaoxian1hao | Hunan | |||
| 35 | Guinongzhan | Guangdong | 89 | 73-1 | Henan | 143 | Xiangzhongxian1hao | Hunan | |||
| 36 | Hongmeizao | Guangdong | 90 | 77xuan7 | Henan | 144 | Yuchi231-8 | Hunan | |||
| 37 | Huanghuazhan | Guangdong | 91 | Dongxuan2hao | Henan | 145 | Aibaiqiu | Fujian | |||
| 38 | Qiguizao25 | Guangdong | 92 | Xinai1hao | Henan | 146 | Aijiaobaimizai | Fujian | |||
| 39 | Qinghuaai6hao | Guangdong | 93 | Aikezao | Zhejiang | 147 | Fushe31 | Fujian | |||
| 40 | Qingliuai | Guangdong | 94 | Aiqing3hao | Zhejiang | 148 | Guangdabai | Fujian | |||
| 41 | Qishanzhan | Guangdong | 95 | Aiwu | Zhejiang | 149 | Hong410 | Fujian | |||
| 42 | Qiuguiai11 | Guangdong | 96 | Butuoai | Zhejiang | 150 | Hongwan52 | Fujian | |||
| 43 | Sanerai | Guangdong | 97 | Erjiuduo | Zhejiang | 151 | Hongyun33 | Fujian | |||
| 44 | Shengai7hao | Guangdong | 98 | Erjiuqing | Zhejiang | 152 | Jinghong1hao | Fujian | |||
| 45 | Shuanggui36 | Guangdong | 99 | Guiluai8hao | Zhejiang | 153 | Longge113 | Fujian | |||
| 46 | Shuangqizhan | Guangdong | 100 | Hongtu31 | Zhejiang | 154 | Lucaihao | Fujian | |||
| 47 | Shuangzhuzhan | Guangdong | 101 | Junxie | Zhejiang | 155 | Minghui63 | Fujian | |||
| 48 | Teqingxuanhui | Guangdong | 102 | Qingganhuang | Zhejiang | 156 | Qiuzhan470 | Fujian | |||
| 49 | Tesanai | Guangdong | 103 | Qingmazao | Zhejiang | 157 | Yuanshannuo | Fujian | |||
| 50 | Texianzhan13 | Guangdong | 104 | Sanjiufeng | Zhejiang | 158 | ZhenD-1 | Fujian | |||
| 51 | Tonghongai | Guangdong | 105 | Shaonuo2hao | Zhejiang | 159 | Zhenhong17 | Fujian | |||
| 52 | Yuexiangzhan | Guangdong | 106 | Shenglong | Zhejiang | 160 | Zhenmu85 | Fujian | |||
| 53 | Zhaiyeqing8hao | Guangdong | 107 | Taizao5hao | Zhejiang | 161 | Zhuliuai | Fujian | |||
| 54 | Zhenguiai1hao | Guangdong | 108 | Wenxuanqing | Zhejiang |
Supplemental Table1 One hundred and sixty-one indica rice varieties from 11 provinces in China.
| No. | Variety | Origin | No. | Variety | Origin | No. | Variety | Origin | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Hucchao1hao | Sichuan | 55 | Zhubao384 | Guangdong | 109 | Xianfeng1hao | Zhejiang | |||
| 2 | Hucheng17 | Sichuan | 56 | Baoxuan2hao | Guangxi | 110 | Zaolian31 | Zhejiang | |||
| 3 | Huhongzao1hao | Sichuan | 57 | Guangnan1hao | Guangxi | 111 | Zaoxian141 | Zhejiang | |||
| 4 | Hunanzao1hao | Sichuan | 58 | Hongnan | Guangxi | 112 | Zhechang3hao | Zhejiang | |||
| 5 | Hushuang1011 | Sichuan | 59 | Jiefang1hao | Guangxi | 113 | Zhengui9hao | Zhejiang | |||
| 6 | Aimakang | Sichuan | 60 | Jinguang202 | Guangxi | 114 | Zhenlong13 | Zhejiang | |||
| 7 | Aituogu151 | Sichuan | 61 | Tuanjie1hao | Guangxi | 115 | Zhenxian97B | Zhejiang | |||
| 8 | CDR22 | Sichuan | 62 | Nanjing1hao | Jiangsu | 116 | Zhongganzao | Zhejiang | |||
| 9 | Chengduai8hao | Sichuan | 63 | Qinglian16 | Jiangsu | 117 | Zhufei10hao | Zhejiang | |||
| 10 | Chenghui448 | Sichuan | 64 | Zhuxi26 | Jiangsu | 118 | Zhuke2hao | Zhejiang | |||
| 11 | Chuanxiang29B | Sichuan | 65 | 508 | Jiangxi | 119 | Zhulianai | Zhejiang | |||
| 12 | Fuhui838 | Sichuan | 66 | 5450 | Jiangxi | 120 | 1296 | Hubei | |||
| 13 | Gang46B | Sichuan | 67 | 6001 | Jiangxi | 121 | Ezao1hao | Hubei | |||
| 14 | K17B | Sichuan | 68 | 6004 | Jiangxi | 122 | Ezao6hao | Hubei | |||
| 15 | Shuhui527 | Sichuan | 69 | 6188 | Jiangxi | 123 | Ezhong2hao | Hubei | |||
| 16 | Yixiang1B | Sichuan | 70 | 7004 | Jiangxi | 124 | Huaai15 | Hubei | |||
| 17 | Annandao | Anhui | 71 | 7005 | Jiangxi | 125 | Jingainuo | Hubei | |||
| 18 | Dengdengqi | Anhui | 72 | Aihong11 | Jiangxi | 126 | Chun389B | Sichuan | |||
| 19 | Mafangxian | Anhui | 73 | Bayiwan | Jiangxi | 127 | Aizizhan | Hunan | |||
| 20 | Wuhu71zao | Anhui | 74 | Gannanzao20 | Jiangxi | 128 | Chenwan3hao | Hunan | |||
| 21 | XieqingzaoB | Anhui | 75 | Gannanzao23 | Jiangxi | 129 | Fanzi | Hunan | |||
| 22 | Zhuguang23 | Anhui | 76 | Jiangxisimiao | Jiangxi | 130 | Hongmidongzhan | Hunan | |||
| 23 | Aijiaonante | Guangdong | 77 | Jingnong3hao | Jiangxi | 131 | Hunanzao | Hunan | |||
| 24 | Conglu51 | Guangdong | 78 | Jingnong69 | Jiangxi | 132 | Laohuanggu | Hunan | |||
| 25 | Fengaizhan1 | Guangdong | 79 | Jiunongzao1hao | Jiangxi | 133 | Liguzao | Hunan | |||
| 26 | Guangchang13 | Guangdong | 80 | Liantangzao | Jiangxi | 134 | Luguiai | Hunan | |||
| 27 | Guangchangai | Guangdong | 81 | Nantehao | Jiangxi | 135 | Xiangaizao10hao | Hunan | |||
| 28 | Guangjie9hao | Guangdong | 82 | Wanxian22 | Jiangxi | 136 | Xiangaizao2hao | Hunan | |||
| 29 | Guangluai4hao | Guangdong | 83 | Xingheng1hao | Jiangxi | 137 | Xiangaizao4hao | Hunan | |||
| 30 | Guangnongai1hao | Guangdong | 84 | Xiujiangwan3hao | Jiangxi | 138 | Xiangaizao7hao | Hunan | |||
| 31 | Guangwen10hao | Guangdong | 85 | Xiujiangzao4hao | Jiangxi | 139 | Xiangaizao8hao | Hunan | |||
| 32 | Guangxuan3hao | Guangdong | 86 | Xiujiangzao9hao | Jiangxi | 140 | Xiangaizao9hao | Hunan | |||
| 33 | Guichao13 | Guangdong | 87 | Youzhanzi | Jiangxi | 141 | Xiangwanxian2hao | Hunan | |||
| 34 | Guichao2hao | Guangdong | 88 | 5006 | Henan | 142 | Xiangzaoxian1hao | Hunan | |||
| 35 | Guinongzhan | Guangdong | 89 | 73-1 | Henan | 143 | Xiangzhongxian1hao | Hunan | |||
| 36 | Hongmeizao | Guangdong | 90 | 77xuan7 | Henan | 144 | Yuchi231-8 | Hunan | |||
| 37 | Huanghuazhan | Guangdong | 91 | Dongxuan2hao | Henan | 145 | Aibaiqiu | Fujian | |||
| 38 | Qiguizao25 | Guangdong | 92 | Xinai1hao | Henan | 146 | Aijiaobaimizai | Fujian | |||
| 39 | Qinghuaai6hao | Guangdong | 93 | Aikezao | Zhejiang | 147 | Fushe31 | Fujian | |||
| 40 | Qingliuai | Guangdong | 94 | Aiqing3hao | Zhejiang | 148 | Guangdabai | Fujian | |||
| 41 | Qishanzhan | Guangdong | 95 | Aiwu | Zhejiang | 149 | Hong410 | Fujian | |||
| 42 | Qiuguiai11 | Guangdong | 96 | Butuoai | Zhejiang | 150 | Hongwan52 | Fujian | |||
| 43 | Sanerai | Guangdong | 97 | Erjiuduo | Zhejiang | 151 | Hongyun33 | Fujian | |||
| 44 | Shengai7hao | Guangdong | 98 | Erjiuqing | Zhejiang | 152 | Jinghong1hao | Fujian | |||
| 45 | Shuanggui36 | Guangdong | 99 | Guiluai8hao | Zhejiang | 153 | Longge113 | Fujian | |||
| 46 | Shuangqizhan | Guangdong | 100 | Hongtu31 | Zhejiang | 154 | Lucaihao | Fujian | |||
| 47 | Shuangzhuzhan | Guangdong | 101 | Junxie | Zhejiang | 155 | Minghui63 | Fujian | |||
| 48 | Teqingxuanhui | Guangdong | 102 | Qingganhuang | Zhejiang | 156 | Qiuzhan470 | Fujian | |||
| 49 | Tesanai | Guangdong | 103 | Qingmazao | Zhejiang | 157 | Yuanshannuo | Fujian | |||
| 50 | Texianzhan13 | Guangdong | 104 | Sanjiufeng | Zhejiang | 158 | ZhenD-1 | Fujian | |||
| 51 | Tonghongai | Guangdong | 105 | Shaonuo2hao | Zhejiang | 159 | Zhenhong17 | Fujian | |||
| 52 | Yuexiangzhan | Guangdong | 106 | Shenglong | Zhejiang | 160 | Zhenmu85 | Fujian | |||
| 53 | Zhaiyeqing8hao | Guangdong | 107 | Taizao5hao | Zhejiang | 161 | Zhuliuai | Fujian | |||
| 54 | Zhenguiai1hao | Guangdong | 108 | Wenxuanqing | Zhejiang |
| Trait | Mean | SD | Max | Min | CV (%) |
|---|---|---|---|---|---|
| GL (mm) | 8.37 | 0.83 | 11.19 | 7.09 | 9.92 |
| GW (mm) | 2.97 | 0.27 | 3.56 | 2.27 | 9.09 |
| TGW (g) | 26.05 | 5.26 | 39.34 | 16.93 | 20.20 |
| GLW | 2.87 | 0.47 | 4.33 | 2.20 | 16.38 |
Table 1 Statistical analysis of grain shape related traits.
| Trait | Mean | SD | Max | Min | CV (%) |
|---|---|---|---|---|---|
| GL (mm) | 8.37 | 0.83 | 11.19 | 7.09 | 9.92 |
| GW (mm) | 2.97 | 0.27 | 3.56 | 2.27 | 9.09 |
| TGW (g) | 26.05 | 5.26 | 39.34 | 16.93 | 20.20 |
| GLW | 2.87 | 0.47 | 4.33 | 2.20 | 16.38 |
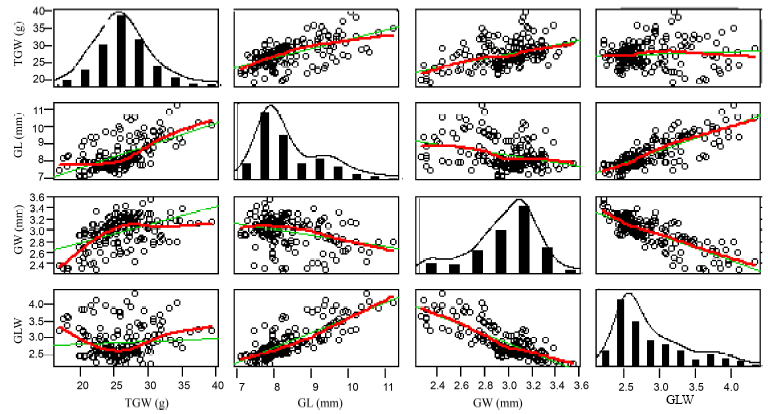
Fig. 1. Statistical analysis of four grain shape traits.TGW, 1000-grain weight; GL, Grain length; GW, Grain width; GLW, Grain length/width.The green lines represent the correlation coefficient, and the red lines represent the correlation trend.
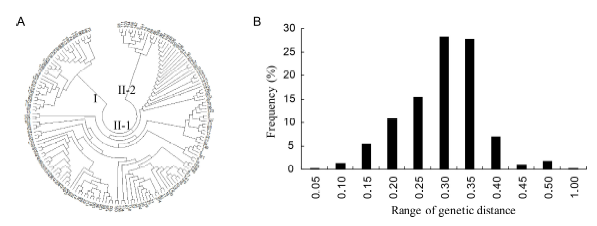
Fig. 2. Population genetic relationship evolution analysis. A, Population structure based on neighbor-joining method. B, Frequency distribution of kinship between varieties.
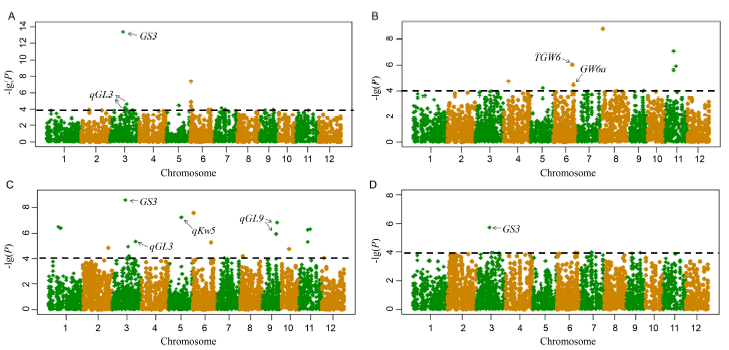
Fig. 3. Manhattan plots for grain length (A), grain width (B), grain length/width (C), and 1000-grain weight (D) by genome-wide association study.The significance threshold -log10(P) is 4.
| Trait | Chr | SNP site a (bp) | Major allele | Minor allele | MAF | -log(P) | Candidate gene/QTL |
|---|---|---|---|---|---|---|---|
| GL | 3 | 17 302 647 | A | G | 0.42 | 13.38 | Os03g0407400 |
| 3 | 19 320 570 | A | G | 0.32 | 4.21 | Os03g0646900 | |
| 3 | 21 821 785 | A | G | 0.33 | 4.60 | Os03g0646900 | |
| 5 | 15 731 341 | A | G | 0.45 | 4.46 | ||
| 6 | 1 764 762 | C | A | 0.10 | 7.38 | ||
| 6 | 1 867 223 | A | G | 0.28 | 4.86 | ||
| 6 | 2 009 738 | C | A | 0.30 | 4.36 | ||
| 7 | 9 004 374 | A | G | 0.43 | 4.11 | ||
| GW | 4 | 6 470 963 | A | G | 0.31 | 4.72 | |
| 5 | 15 731 341 | A | G | 0.45 | 4.23 | ||
| 6 | 24 170 016 | C | A | 0.49 | 6.03 | Os06g0623700 | |
| 6 | 26 025 122 | A | G | 0.47 | 4.45 | Os06g0650300 | |
| 8 | 3 681 009 | A | G | 0.27 | 8.80 | ||
| 9 | 20 143 516 | A | G | 0.37 | 4.01 | ||
| 11 | 11 962 160 | G | A | 0.24 | 5.61 | ||
| 11 | 12 028 646 | G | A | 0.19 | 7.08 | ||
| 11 | 14 891 499 | A | T | 0.20 | 5.93 | ||
| GLW | 1 | 12 869 918 | A | T | 0.19 | 6.49 | |
| 1 | 15 765 545 | A | G | 0.26 | 6.40 | ||
| 2 | 31 232 596 | A | C | 0.49 | 4.86 | ||
| 3 | 17 302 647 | A | G | 0.42 | 8.61 | Os03g0407400 | |
| 3 | 19 320 570 | A | G | 0.32 | 4.05 | ||
| 3 | 20 622 588 | A | C | 0.15 | 4.95 | Os03g0646900 | |
| 3 | 21 821 785 | A | G | 0.33 | 4.19 | ||
| 3 | 30 021 901 | A | G | 0.42 | 5.33 | ||
| 5 | 15 731 341 | A | G | 0.45 | 7.24 | qKw5 | |
| 6 | 1 764 762 | C | A | 0.10 | 7.58 | ||
| 6 | 24 170 016 | C | A | 0.49 | 5.26 | ||
| 7 | 9 004 374 | A | G | 0.43 | 4.05 | ||
| 8 | 3 681 009 | A | G | 0.27 | 4.19 | ||
| 9 | 19 020 266 | A | C | 0.41 | 5.93 | qGL9 | |
| 9 | 20 143 516 | A | G | 0.37 | 6.83 | qGL9 | |
| 10 | 11 961 180 | T | A | 0.43 | 4.74 | ||
| 11 | 11 962 160 | G | A | 0.24 | 5.30 | ||
| 11 | 12 028 646 | G | A | 0.19 | 6.27 | ||
| 11 | 14 891 499 | A | T | 0.20 | 6.31 | ||
| 12 | 2 963 765 | G | A | 0.34 | 4.06 | ||
| TGW | 3 | 17 302 647 | A | G | 0.42 | 5.72 | Os03g0407400 |
Table 2 Thirty-eight single nucleotide polymorphisms (SNPs) significantly associated with grain size by genome-wide association study.
| Trait | Chr | SNP site a (bp) | Major allele | Minor allele | MAF | -log(P) | Candidate gene/QTL |
|---|---|---|---|---|---|---|---|
| GL | 3 | 17 302 647 | A | G | 0.42 | 13.38 | Os03g0407400 |
| 3 | 19 320 570 | A | G | 0.32 | 4.21 | Os03g0646900 | |
| 3 | 21 821 785 | A | G | 0.33 | 4.60 | Os03g0646900 | |
| 5 | 15 731 341 | A | G | 0.45 | 4.46 | ||
| 6 | 1 764 762 | C | A | 0.10 | 7.38 | ||
| 6 | 1 867 223 | A | G | 0.28 | 4.86 | ||
| 6 | 2 009 738 | C | A | 0.30 | 4.36 | ||
| 7 | 9 004 374 | A | G | 0.43 | 4.11 | ||
| GW | 4 | 6 470 963 | A | G | 0.31 | 4.72 | |
| 5 | 15 731 341 | A | G | 0.45 | 4.23 | ||
| 6 | 24 170 016 | C | A | 0.49 | 6.03 | Os06g0623700 | |
| 6 | 26 025 122 | A | G | 0.47 | 4.45 | Os06g0650300 | |
| 8 | 3 681 009 | A | G | 0.27 | 8.80 | ||
| 9 | 20 143 516 | A | G | 0.37 | 4.01 | ||
| 11 | 11 962 160 | G | A | 0.24 | 5.61 | ||
| 11 | 12 028 646 | G | A | 0.19 | 7.08 | ||
| 11 | 14 891 499 | A | T | 0.20 | 5.93 | ||
| GLW | 1 | 12 869 918 | A | T | 0.19 | 6.49 | |
| 1 | 15 765 545 | A | G | 0.26 | 6.40 | ||
| 2 | 31 232 596 | A | C | 0.49 | 4.86 | ||
| 3 | 17 302 647 | A | G | 0.42 | 8.61 | Os03g0407400 | |
| 3 | 19 320 570 | A | G | 0.32 | 4.05 | ||
| 3 | 20 622 588 | A | C | 0.15 | 4.95 | Os03g0646900 | |
| 3 | 21 821 785 | A | G | 0.33 | 4.19 | ||
| 3 | 30 021 901 | A | G | 0.42 | 5.33 | ||
| 5 | 15 731 341 | A | G | 0.45 | 7.24 | qKw5 | |
| 6 | 1 764 762 | C | A | 0.10 | 7.58 | ||
| 6 | 24 170 016 | C | A | 0.49 | 5.26 | ||
| 7 | 9 004 374 | A | G | 0.43 | 4.05 | ||
| 8 | 3 681 009 | A | G | 0.27 | 4.19 | ||
| 9 | 19 020 266 | A | C | 0.41 | 5.93 | qGL9 | |
| 9 | 20 143 516 | A | G | 0.37 | 6.83 | qGL9 | |
| 10 | 11 961 180 | T | A | 0.43 | 4.74 | ||
| 11 | 11 962 160 | G | A | 0.24 | 5.30 | ||
| 11 | 12 028 646 | G | A | 0.19 | 6.27 | ||
| 11 | 14 891 499 | A | T | 0.20 | 6.31 | ||
| 12 | 2 963 765 | G | A | 0.34 | 4.06 | ||
| TGW | 3 | 17 302 647 | A | G | 0.42 | 5.72 | Os03g0407400 |
| Number | Variety | GL (mm) | GW (mm) | GS3 haplotype | TGW6 haplotype |
|---|---|---|---|---|---|
| 1 | Aijiaonante | 7.94 | 3.40 | GS3-12 | TGW6-1 |
| 2 | Guangluai4 | 7.49 | 3.56 | GS3-7 | TGW6-1 |
| 3 | Nantehao | 7.74 | 3.21 | GS3-2 | TGW6-1 |
| 4 | Guizhao2 | 6.99 | 3.33 | GS3-3 | TGW6-2 |
| 5 | Chenwan3 | 7.18 | 3.10 | GS3-4 | TGW6-1 |
| 6 | Aimakang | 7.47 | 2.98 | GS3-5 | TGW6-1 |
| 7 | Aizizhan | 8.01 | 2.75 | GS3-4 | TGW6-5 |
| 8 | Teqingxuanhui | 8.91 | 3.10 | GS3-1 | TGW6-3 |
| 9 | G Zhenshan97B | 9.36 | 2.84 | GS3-6 | TGW6-1 |
| 10 | Xiangaizao10hao | 7.80 | 3.30 | GS3-6 | TGW6-3 |
| 11 | Aituogu151 | 6.98 | 3.36 | GS3-13 | TGW6-1 |
| 12 | Gang46B | 7.68 | 3.30 | GS3-9 | TGW6-1 |
| 13 | Guangluai4 | 7.44 | 3.54 | GS3-10 | TGW6-1 |
| 14 | Minghui63 | 9.62 | 2.84 | GS3-11 | TGW6-5 |
| 15 | Yuexiangzhan | 7.66 | 2.54 | GS3-10 | TGW6-5 |
| 16 | Jiangxisimiao | 9.28 | 2.64 | GS3-8 | TGW6-5 |
| 17 | Chenghui448 | 9.02 | 2.80 | GS3-11 | TGW6-3 |
| 18 | Fengaizhan | 8.28 | 2.41 | GS3-14 | TGW6-5 |
| 19 | Annanzao | 8.75 | 2.87 | GS3-5 | TGW6-1 |
| 20 | Dengdengqi | 7.68 | 2.90 | GS3-5 | TGW6-1 |
| 21 | Tesanai | 7.51 | 3.31 | GS3-5 | TGW6-5 |
| 22 | Huanghuazhan | 8.30 | 2.67 | GS3-4 | TGW6-4 |
Supplemental Table 2 Phenotype and genotype classification of 22 rice varieties.
| Number | Variety | GL (mm) | GW (mm) | GS3 haplotype | TGW6 haplotype |
|---|---|---|---|---|---|
| 1 | Aijiaonante | 7.94 | 3.40 | GS3-12 | TGW6-1 |
| 2 | Guangluai4 | 7.49 | 3.56 | GS3-7 | TGW6-1 |
| 3 | Nantehao | 7.74 | 3.21 | GS3-2 | TGW6-1 |
| 4 | Guizhao2 | 6.99 | 3.33 | GS3-3 | TGW6-2 |
| 5 | Chenwan3 | 7.18 | 3.10 | GS3-4 | TGW6-1 |
| 6 | Aimakang | 7.47 | 2.98 | GS3-5 | TGW6-1 |
| 7 | Aizizhan | 8.01 | 2.75 | GS3-4 | TGW6-5 |
| 8 | Teqingxuanhui | 8.91 | 3.10 | GS3-1 | TGW6-3 |
| 9 | G Zhenshan97B | 9.36 | 2.84 | GS3-6 | TGW6-1 |
| 10 | Xiangaizao10hao | 7.80 | 3.30 | GS3-6 | TGW6-3 |
| 11 | Aituogu151 | 6.98 | 3.36 | GS3-13 | TGW6-1 |
| 12 | Gang46B | 7.68 | 3.30 | GS3-9 | TGW6-1 |
| 13 | Guangluai4 | 7.44 | 3.54 | GS3-10 | TGW6-1 |
| 14 | Minghui63 | 9.62 | 2.84 | GS3-11 | TGW6-5 |
| 15 | Yuexiangzhan | 7.66 | 2.54 | GS3-10 | TGW6-5 |
| 16 | Jiangxisimiao | 9.28 | 2.64 | GS3-8 | TGW6-5 |
| 17 | Chenghui448 | 9.02 | 2.80 | GS3-11 | TGW6-3 |
| 18 | Fengaizhan | 8.28 | 2.41 | GS3-14 | TGW6-5 |
| 19 | Annanzao | 8.75 | 2.87 | GS3-5 | TGW6-1 |
| 20 | Dengdengqi | 7.68 | 2.90 | GS3-5 | TGW6-1 |
| 21 | Tesanai | 7.51 | 3.31 | GS3-5 | TGW6-5 |
| 22 | Huanghuazhan | 8.30 | 2.67 | GS3-4 | TGW6-4 |
| [1] | Fan C C, Xing Y Z, Mao H L, Lu T T, Han B, Xu C G, Li X H, Zhang Q F.2006. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet, 112(6): 1164-1171. |
| [2] | Fan C C, Yu S B, Wang C R, Xing Y Z.2009. A causal C-A mutation in the second exon of GS3 highly associated with rice grain length and validated as a functional marker. Theor Appl Genet, 118(3): 465-472. |
| [3] | Fu F H, Wang F.1994. Genetic analysis of grain characters in hybrid rice. Acta Agron Sin, 20(1): 39-45. (in Chinese with English abstract) |
| [4] | Gao X Y, Zhang X J, Lan H X, Huang J, Wang J F, Zhang H S.2015. The additive effects of GS3 and qGL3 on rice grain length regulation revealed by genetic and transcriptome comparisons. BMC Plant Biol, 15(1): 156. |
| [5] | Gao Z Q, Zhan X D, Liang Y S, Cheng S H, Cao L Y.2011. Research advances on the inheritance of rice grain shape traits and related gene mapping and cloning.Genetics, 33(4): 314-321. (in Chinese with English abstract) |
| [6] | Glaubitz J C.2010. CONVERT: A user-friendly program to reformat diploid genotypic data for commonly used population genetic software packages. Mol Ecol Resour, 4(2): 309-310. |
| [7] | Gong L H, Gao Z Y, Ma B J, Qian Q.2011. Advances in research on rice grain size genetics.Chin Bull Bot, 46(6): 597-605. (in Chinese with English abstract) |
| [8] | Guo L B, Ye G Y.2014. Use of major quantitative trait loci to improve grain yield of rice.Rice Sci, 21(2): 65-82. |
| [9] | Hu J, Wang Y X, Fang Y X, Zeng L J, Xu J, Yu H P, Shi Z Y, Pan J J, Zhang D, Kang S J, Zhu L, Dong D J, Guo L B, Zeng D L, Zhang G H, Xie L H, Xiong G S, Li J Y, Qian Q.2015. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant, 8(10): 1455-1465. |
| [10] | Huang H X, Qian Q.2017. Progress in genetic research of rice grain shape and breeding achievements of long-grain shape and good qualityjaponica rice. Chin J Rice Sci, 31(6): 665-672. (in Chinese with English abstract) |
| [11] | Huang X H, Wei X H, Sang T, Zhao Q, Feng Q, Zhao Y, Li C Y, Zhu C R, Lu T T, Zhang Z W, Li M, Fan D L, Guo Y L, Wang A H, Wang L, Deng L W, Li W J, Lu Y Q, Weng Q J, Liu K Y, Huang T, Zhou T Y, Jing Y F, Li W, Lin Z, Buckler E S, Qian Q, Zhang Q F, Li J Y, Han B.2010. Genome-wide association studies of 14 agronomic traits in rice landraces.Nat Genet, 42: 961-967. |
| [12] | Huang X H, Wei X H, Sang T, Zhao Q, Feng Q, Zhao Y, Li C Y, Zhu C R, Lu T T, Zhang Z W, Li M, Fan D L, Guo Y L, Wang A H, Wang L, Deng L W, Li W J, Lu Y Q, Weng Q J, Liu K Y, Huang T, Zhou T Y, Jing Y F, Li W, Zhang L, Qian Q, Zhang Q F, Li J Y.2012. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm.Nat Genet, 44: 32-39. |
| [13] | Huang X H, Han B.2014. Natural variations and genome-wide association studies in crop plants.Ann Rev Plant Biol, 65(1): 531-551. |
| [14] | Ishimaru K, Hirotsu N, Madoka Y, Murakami N, Hara N, Onodera H, Kashiwagi T, Ujiie K, Shimizu B, Onishi A, Miyagawa H, Katoh E.2013. Loss of function of the IAA-glucose hydrolase geneTGW6 enhances rice grain weight and increases yield. Nat Genet, 45(6): 707-711. |
| [15] | Lang Y L.2015. A preliminary analysis of genetic effects of seven rice genotypes [MS Thesis]. Nanjing: Nanjing Agricultural University. (in Chinese with English abstract) |
| [16] | Li M M, Xu L, Liu C W, Cao G L, He H H, Han L Z.2008. Advances in rice grain shape inheritance and QTLs mapping.China Agric Technol Rev, 10(1): 34-42. (in Chinese with English abstract) |
| [17] | Li Y B, Fan C C, Xing Y Z, Jiang Y H, Luo L J, Sun L, Shao D, Xu L J, Li X H, Xiao J H, He Y Q, Zhang Q F.2011. Natural variation inGS5 plays an important role in regulating grain size and yield in rice. Nat Genet, 43(12): 1266-1269. |
| [18] | Lin L H, Wu W R.2003. QTL mapping of grain shape and grain weight in rice.Mol Plant Breeding, 1(3): 337-342. (in Chinese with English abstract) |
| [19] | Liu K, Muse S V.2005. PowerMarker: An integrated analysis environment for genetic marker analysis.Bioinformatics, 21(9): 2128-2129. |
| [20] | Liu X, Mou C L, Zhou C L, Cheng Z J, Jiang L, Wan J M.2018. Research progress on cloning and regulation mechanism of rice grain shape genes.Chin J Rice Sci, 32(1): 1-11. (in Chinese with English abstract) |
| [21] | Mao H L, Sun S Y, Yao J L, Wang C R, Yu S B, Xu C Q, Li X H, Zhang Q F.2010. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc Natl Acad Sci USA, 107: 19579-19584. |
| [22] | Prasad K, Parameswaran S, Vijayraghavan U.2010. OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an earlyacting regulator of inner floral organs. Plant J, 43(6): 915-928. |
| [23] | Qi P, Lin Y S, Song X J, Shen J B, Huang W, Shan J X, Zhu M Z, Jiang L, Gao J P, Lin H X.2012. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1: 3. Cell Res, 22(12): 1666-1680. |
| [24] | Qiu X J.2013. Identification and identification of a major gene qSS7 in rice grain. [PhD Thesis]. Wuhan: Huazhong Agricultural University. (in Chinese with English abstract) |
| [25] | Rabiei B, Valizadeh M, Ghareyazie B, Moghaddam M, Ali A J.2004. Identification of QTLs for rice grain size and shape of Iranian cultivars using SSR markers.Euphytica, 137(3): 325-332. |
| [26] | Redoña E D, Mackill D J.1998. Quantitative trait locus analysis for rice panicle and grain characteristics.Theor Appl Genet, 96: 957-963. |
| [27] | Risch N, Merikangas K.1996. The future of genetic studies of complex human diseases.Science, 273: 1516-1517. |
| [28] | Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K Saito T, Kobayashi M, Khush G S, Kitano H, Matsuoka M.2002. Green revolution: A mutant gibberellin-synthesis gene in rice. Nature, 416: 701-702. |
| [29] | Shi C H, Shen Z T.1995. Genetics and improvement of early grain shape.Chin J Rice Sci, 9(1): 27-32. (in Chinese with English abstract) |
| [30] | Si L Z, Chen J Y, Huang X H, Gong H, Luo J H, Hou Q Q, Zhou T Y, Lu T T, Zhu J J, Shang G Y, Chen E W, Gong C X, Zhao Q, Jing Y F, Zhao Y, Li Y, Cui L L, Fan D L, Lu Y Q, Weng Q J, Wang Y C, Zhan Q L, Liu K Y, Wei X H, An K, An G, Han B.2016. OsSPL13 controls grain size in cultivated rice. Nat Genet, 48: 447-456. |
| [31] | Song X J, Huang W, Shi M, Zhu M Z, Lin H X.2007. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet, 39(5): 623-630. |
| [32] | Wan X Y, Weng J F, Zhai H Q, Wang J K, Lei C L, Liu X L, Guo T, Jiang L, Su N, Wan J M.2008. Quantitative trait loci (QTL) analysis for rice grain width and fine mapping of an identified QTL allele gw-5 in a recombination hotspot region on chromosome 5. Genetics, 179(4): 2239-2252. |
| [33] | Wang S K, Wu K, Yuan Q B, Liu X Y, Liu Z B, Lin X Y, Zeng R Z, Zhu H T, Dong G J, Qian Q, Zhang G Q, Fu X D.2012. Control of grain size, shape and quality byOsSPL16 in rice. Nat Genet, 44(8): 950-954. |
| [34] | Wang Y X, Xiong G S, Hu J, Jiang L, Yu H, Xu J, Fang Y X, Zeng L J, Xu E B, Xu J, Ye W J, Meng X B, Liu R F, Chen H Q, Jing Y H, Wang Y H, Zhu X D, Li J Y, Qian Q.2015. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat Genet, 47(8): 944-948. |
| [35] | Wei X H, Yuan X P, Yu H Y, Wang Y P, Xu Q, Tang S X.2009. Analysis of genetic variation of main rice varieties in China.Chin J Rice Sci, 23(3): 237-244. (in Chinese with English abstract) |
| [36] | Wright M H, Tung C W, Zhao K, Reynolds A, McCouch S R, Bustamante C D.2010. ALCHEMY: A reliable method for automated SNP genotype calling for small batch sizes and highly homozygous populations.Bioinformatics, 26(23): 2952-2960. |
| [37] | Wu C M, Sun C Q, Chen L, Li Z C, Wang X K.2002. Analysis QTL of grain shape by using of RFLP map in rice.Jilin Agric Sci, 27(5): 3-7. (in Chinese with English abstract) |
| [38] | Xuan Y S, Jiang W S, Liu X H, Cheng H Z, Hee-Jong Koh, Yuan D L.2010. Genetic diversity of main rice cultivars in northeast China.J Plant Genet Res, 11(2): 206-212. (in Chinese with English abstract) |
| [39] | Yang L S, Bai Y S, Xu C W, Hu X M, Wang W M, She D H, Chen G Z.2001. Advances in research on rice grain shape and its inheritance.Anhui Agric Sci, 29(2): 164-167. (in Chinese with English abstract) |
| [40] | Yang T F, Zeng R Z, Zhu H T.2010. Effect of rice grain length geneGS3 on polymerization breeding. Mol Plant Breeding, 8(1): 59-66. |
| [41] | Yano K, Yamamoto E, Aya K, Takeuchi H, Lo P C, Hu L, Yamasaki M, Yoshida S, Kitano H, Hirano K, Matsuoka M.2016. Genome- wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice.Nat Genet, 48(8): 927-934. |
| [42] | Zhang G H, Zhang G P, Qian Q, Xu L P, Zeng D L, Teng S, Bao J S.2004. Analysis of QTLs for quantitative traits of rice grain under different environmental conditions.Chin J Rice Sci, 18(1): 16-22. (in Chinese with English abstract) |
| [43] | Zhao D S, Li Q F, Zhang C Q, Zhang C, Yang Q Q, Pan L X, Ren X Y, Lu J, Gu M H, Liu Q Q.2018. GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat Commun, 9(1): 1240. |
| [44] | Zhao K Y, Tung C W, Eizenga G C, Wright M H, Ali L M, Price A H, Norton G J, Islam M R, Reynolds A, Mezey J, McClung A M, Bustamante C D, McCouch S R.2011. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat Commun, 2(1): 467. |
| [45] | Zhu L H.2007. My opinion about high yield rice breeding in China.J Nanjing Agric Univ, 30(1): 129-135. (in Chinese with English abstract) |
| [46] | Zuo S M, Kang H X, Li Q Q, Chen Z X, Zhang Y F, Liu W D, Wang G L, Chen H Q, Pan X B.2014. Genome-wide association analysis and utilization of gene related to ear traits in introduced rice germplasm.Chin J Rice Sci, 28(6): 649-658. (in Chinese with English abstract). |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [13] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [14] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [15] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||