
Rice Science ›› 2021, Vol. 28 ›› Issue (6): 557-566.DOI: 10.1016/j.rsci.2021.05.014
• Research Paper • Previous Articles Next Articles
Tao Wang, Lijuan Lou, Zeyu Li, Lianguang Shang, Quan Wang( )
)
Received:2020-08-15
Accepted:2021-05-14
Online:2021-11-28
Published:2021-11-28
Tao Wang, Lijuan Lou, Zeyu Li, Lianguang Shang, Quan Wang. Cloning and Characterization of Protein Prenyltransferase Alpha Subunit in Rice[J]. Rice Science, 2021, 28(6): 557-566.
Add to citation manager EndNote|Ris|BibTeX
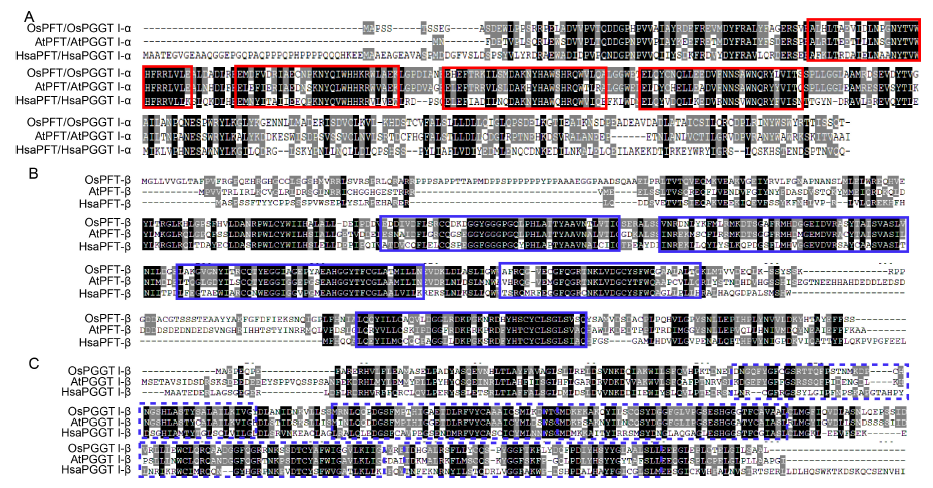
Fig. 1. Multiple sequence alignment of deduced amino acid sequences of prenyltransferase subunit and molecular.A, Amino acid sequence alignment of OsPFT/OsPGGT I-α subunit with HsaPFT/HsaPGGT I-α and AtPFT/AtPGGT I-α subunits. Letters in the black boxes indicate highly conserved amino acid residues. The red solid frames represent the protein prenyltransferase alpha subunit repeat domain.B, Amino acid sequence alignment of OsPFT-β subunit with HsaPFT-β and AtPFT-β subunits. The blue solid frames represent the prenyltransferase and squalene oxidase repeat domain.C, Amino acid sequence alignment of OsPGGT I-β subunit with HsaPGGT I-β and AtPGGT I-β subunits. The blue dotted frames represent the prenyltransferase and squalene oxidase repeat domain.
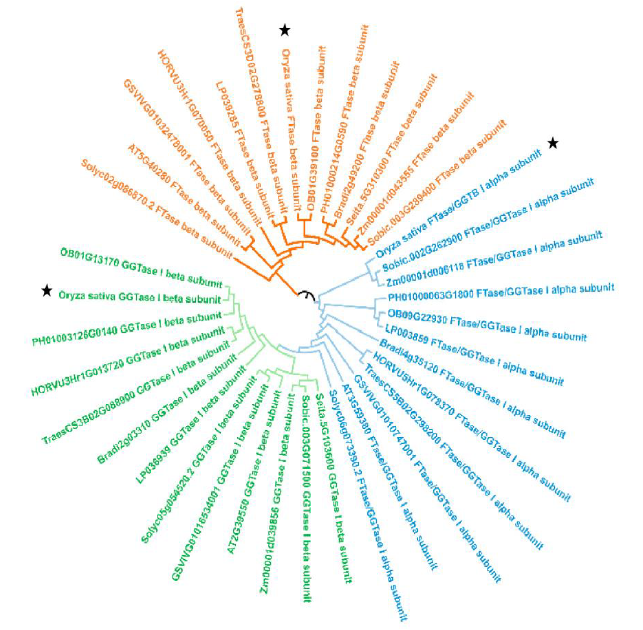
Fig. S2. Molecular phylogenetic tree analysis of selected prenyltransferase subunit amino acids among different species.The construction of phylogenetic tree is based on the maximum likelihood method in MEGA7. Bootstrap value = 1000. The pentagram indicates the corresponding prenyltransferase subunit in rice.
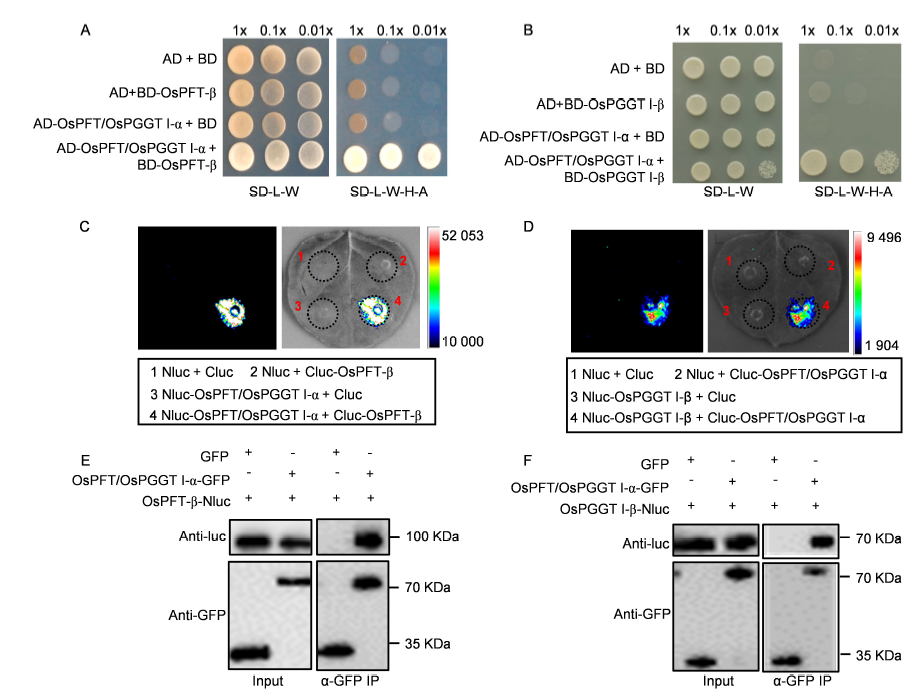
Fig. 2. OsPFT/OsPGGT I-α interacts with OsPFT-β and OsPGGT I-β.A and B, Yeast-two hybrid analysis of OsPFT/OsPGGT I-α and OsPFT-β interaction (A) and OsPFT/PGGT I-α and OsPGGT I-β interaction (B). Bait of OsPFT-β or OsPGGT I-β was fused to the GAL4 DNA-binding domain, and prey of OsPFT/OsPGGT I-α was fused to the GAL4 activation domain. Colonies growth on the SD-L-W-H-A medium represent positive interaction.C and D, Firefly luciferase complementation imaging (LCI) assay in Nicotiana benthamiana. The N-terminal half of luciferase (Nluc) was fused into OsPFT/OsPGGT I-α or OsPGGT I-β and the C-terminal half of luciferase (Cluc) was fuse into OsPFT/OsPGGT I-α or OsPFT-β.E and F, Co-immunoprecipitation (Co-IP) assay for OsPFT/OsPGGT I-α and OsPFT-β (E) or OsPFT/OsPGGT I-α and OsPGGT I-β (F). OsPFT/OsPGGT I-α was fused to green fluorescent protein (GFP), OsPFT-β and OsPGGT I-β were fused to Nluc, respectively. These constructions and GFP control vector were transformed into Agrobacterium GV3101 strain and infiltrated into N. benthamiana. OsPFT/OsPGGT I-α-GFP or GFP were immobilized on GFP-trap beads and incubated with OsPFT-β-Nluc or OsPGGT I-β-Nluc. Input represents equal amounts of purified OsPFT/OsPGGT I-α-GFP or GFP fusions and OsPFT-β-Nluc or OsPGGT I-β-Nluc before Co-IP.
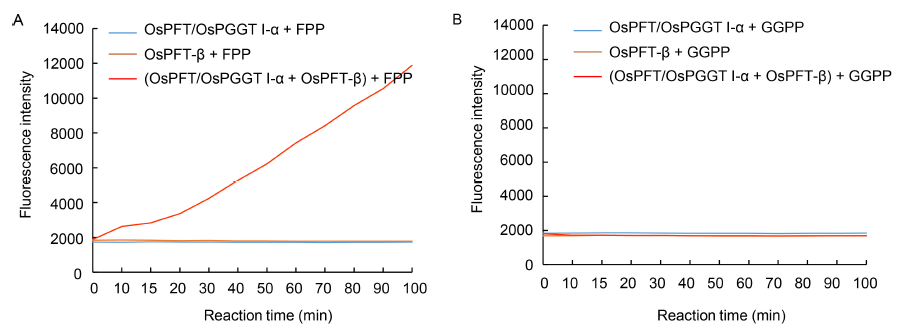
Fig. 3. In vitro continuous ?uorescence assay for OsPFT/OsPGGT I-α and OsPFT-β activity measurement.A, In vitro OsPFT/OsPGGT I-α and OsPFT-β activity assay using farnesyl pyrophosphate (FPP) as substrate. OsPFT/OsPGGT I-α + FPP and OsPFT-β + FPP were used as the negative controls.B, In vitro OsPFT/OsPGGTI-α and OsPFT-β activity assay using geranyl geranyl pyrophosphate (GGPP) as substrate. OsPFT/OsPGGT I-α + GGPP and OsPFT-β + GGPP were used as the negative controls.The fluorescence assays were performed in the Tecan Infinite Pro Microplate Reader and monitored using an excitation wavelength of 340 nm and emission wavelength of 505 nm.
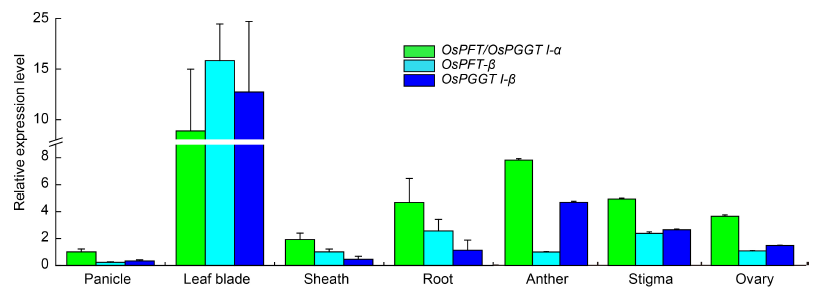
Fig. 4. Gene expression pattern in different orgens and tissues.Gene expression levels of OsPFT/OsPGGT I-α, OsPFT-β and OsPGGT I-β in different rice organs or tissues. Rice Actin was used as the reference. Data are Mean ± SD (n = 3).
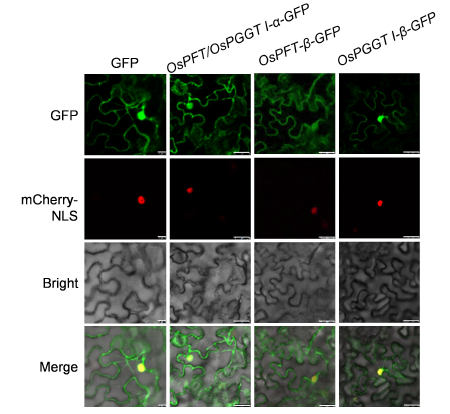
Fig. 5. Subcellular localization assay of GFP-tagging OsPFT/OsPGGT I-α, OsPFT-β and OsPGGT I-β in Nicotiana benthamiana.The assays were performed using mCherry-NLS as a nucleus-location signal by the confocal laser microscope scanning. The scale bar is 10 μm in the first column, and 25 μm in the next three columns.

Fig. 6. Mutation analysis in OsPFT/OsPGGT I-α by CRISPR/Cas9-mediated genome editing system.A, Schematic diagram of the designed targeted sites in OsPFT/OsPGGT I-α. 3′/5′ UTRs, exons and introns are indicated by green rectangles, blue rectangles and black lines, respectively. The target sequences are underlined, and the protospacer adjacent motif (PAM) is emphasized in red letters. Sequence analyses of the targeted regions in OsPFT/OsPGGT I-α are in T3 transgenic plants. The deletion sequence is shown by short solid line. CR-1 and CR-2 refer to the two OsPFT/OsPGGT I-α mutants; WT refers to the wild type.B, PCR assay of T3 transgenic lines in CR-2. Each lane represents an independent CRISPR/Cas-mediated editing line.C, Comparison of WT and Hetero CR-1/CR-2 in floret at the mature pollen stage. Hetero CR-1 and Hetero CR-2 represent the heterozygous OsPFT/OsPGGT I-αCR-1/2. Scale bars are 1 mm.D, Pollen was stained with I2-KI. The red arrows marked the sterile pollen. Scale bars are 30 μm.E, Statistic of sterile pollen. The error bars at each data point indicate the standard error obtained from three biological replicates. **, Significant difference at the 0.01 level.
| [1] | Allen G J, Murata Y, Chu S P, Nafisi M, Schroeder J I. 2002. Hypersensitivity of abscisic acid-induced cytosolic calcium increases in the Arabidopsis farnesyltransferase mutant era1-2. Plant Cell, 14(7): 1649-1662. |
| [2] | Barghetti A, Sjogren L, Floris M, Paredes E B, Wenkel S, Brodersen P. 2017. Heat-shock protein 40 is the key farnesylation target in meristem size control, abscisic acid signaling, and drought resistance. Gene Dev, 31(22): 2282-2295. |
| [3] | Bonetta D, Bayliss P, Sun S, Sage T, McCourt P. 2000. Farnesylation is involved in meristem organization in Arabidopsis. Planta, 211(2): 182-190. |
| [4] | Brady S M, Sarkar S F, Bonetta D, McCourt P. 2003. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J, 34(1): 67-75. |
| [5] | Chen H M, Zou Y, Shang Y L, Lin H Q, Wang Y J, Cai R, Tang X Y, Zhou J M. 2008. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol, 146(2): 368-376. |
| [6] | Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. 1996. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science, 273: 1239-1241. |
| [7] | Daszkowska-Golec A, Skubacz A, Sitko K, Slota M, Kurowska M, Szarejko I. 2018. Mutation in barley ERA1 (Enhanced Response to ABA1) gene confers better photosynthesis efficiency in response to drought as revealed by transcriptomic and physiological analysis. Environ Exp Bot, 148: 12-26. |
| [8] | Defeojones D, Tatchell K, Robinson L C, Sigal I S, Vass W C, Lowy D R, Scolnick E M. 1985. Mammalian and yeast ras gene products: Biological function in their heterologous systems. Science, 228: 179-184. |
| [9] | Dozier J K, Khatwani S L, Wollack J W, Wang Y C, Schmidt-Dannert C, Distefano M D. 2014. Engineering protein farnesyltransferase for enzymatic protein labeling applications. Bioconjugate Chem, 25(7): 1203-1212. |
| [10] | Hala M, Elias M, Zarsky V. 2005. A specific feature of the angiosperm Rab escort protein (REP) and evolution of the REP/GDI superfamily. J Mol Biol, 348(5): 1299-1313. |
| [11] | Kamiya Y, Sakurai A, Tamura S, Takahashi N, Abe K, Tsuchiya E, Fukui S, Kitada C, Fujino M. 1978. Structure of rhodotorucine A, a novel lipopeptide, inducing mating tube formation in Rhodosporidium toruloides. Biochem Biophys Res Commun, 83(3): 1077-1083. |
| [12] | Kataoka T, Powers S, Cameron S, Fasano O, Goldfarb M, Broach J, Wigler M. 1985. Functional homology of mammalian and yeast RAS genes. Cell, 40(1): 19-26. |
| [13] | Leung K F, Baron R, Seabra M C. 2006. Thematic review series: Lipid posttranslational modifications. Geranylgeranylation of Rab GTPases. J Lipid Res, 47(3): 467-475. |
| [14] | Manmathan H, Shaner D, Snelling J, Tisserat N, Lapitan N. 2013. Virus-induced gene silencing of Arabidopsis thaliana gene homologues in wheat identifies genes conferring improved drought tolerance. J Exp Bot, 64(5): 1381-1392. |
| [15] | Maurer-Stroh S, Washietl S, Eisenhaber F. 2003. Protein prenyltransferases. Genome Biol, 4(4): 212. |
| [16] | Northey J G B, Liang S Y, Jamshed M, Deb S, Foo E, Reid J B, McCourt P, Samuel M A. 2016. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat Plants, 2(8): 16114. |
| [17] | Ogata T, Nagatoshi Y, Yamagishi N, Yoshikawa N, Fujita Y. 2017. Virus-induced down-regulation of GmERA1A and GmERA1B genes enhances the stomatal response to abscisic acid and drought resistance in soybean. PLoS One, 12(4): e0175650. |
| [18] | Pei Z M, Ghassemian M, Kwak C M, McCourt P, Schroeder J I. 1998. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science, 282: 287-290. |
| [19] | Rowinsky E K, Windle J J, Von Hoff D D. 1999. Ras protein farnesyltransferase: A strategic target for anticancer therapeutic development. J Clin Oncol, 17(11): 3631-3652. |
| [20] | Running M P, Lavy M, Sternberg H, Galichet A, Gruissem W, Hake S, Ori N, Yalovsky S. 2004. Enlarged meristems and delayed growth in plp mutants result from lack of CaaX prenyltransferases. Proc Natl Acad Sci USA, 101(20): 7815-7820. |
| [21] | Running M P. 2014. The role of lipid post-translational modification in plant developmental processes. Front Plant Sci, 5: 50. |
| [22] | Sakagami Y, Yoshida M, Isogai A, Suzuki A. 1981. Peptidal sex-hormones inducing conjugation tube formation in compatible mating-type cells of Tremella mesenterica. Science, 212: 1525- 1527. |
| [23] | Thole J M, Perroud P F, Quatrano R S, Running M P. 2014. Prenylation is required for polar cell elongation, cell adhesion, and differentiation in Physcomitrella patens. Plant J, 78(3): 441-451. |
| [24] | Turnbull D, Hemsley P A. 2017. Fats and function: Protein lipid modifications in plant cell signalling. Curr Opin Plant Biol, 40: 63-70. |
| [25] | Wang Y, Ying J F, Kuzma M, Chalifoux M, Sample A, McArthur C, Uchacz T, Sarvas C, Wan J X, Dennis D T, McCourt P, Huang Y F. 2005. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J, 43(3): 413-424. |
| [26] | Wang Y, Beaith M, Chalifoux M, Ying J F, Uchacz T, Sarvas C, Griffiths R, Kuzma M, Wan J X, Huang Y F. 2009. Shoot- specific down-regulation of protein farnesyltransferase (alpha- subunit) for yield protection against drought in Canola. Mol Plant, 2(1): 191-200. |
| [27] | Wu J R, Wang T Y, Weng C P, Duong N K T, Wu S J. 2019. AtJ3, a specific HSP40 protein, mediates protein farnesylation- dependent response to heat stress in Arabidopsis. Planta, 250(5): 1449-1460. |
| [28] | Yalovsky S, Rodr G M, Gruissem W. 1999. Lipid modifications of proteins-slipping in and out of membranes. Trends Plant Sci, 4(11): 439-445. |
| [29] | Yalovsky S, Kulukian A, Rodriguez-Concepcion M, Young C A, Gruissem W. 2000a. Functional requirement of plant farnesyltransferase during development in Arabidopsis. Plant Cell, 12(8): 1267-1278. |
| [30] | Yalovsky S, Rodriguez-Concepcion M, Bracha K, Toledo-Ortiz G, Gruissem W. 2000b. Prenylation of the floral transcription factor APETALA1 modulates its function. Plant Cell, 12(8): 1257-1266. |
| [31] | Zhang F L, Casey P J. 1996. Protein prenylation: Molecular mechanisms and functional consequences. Annu Rev Biochem, 65: 241-269. |
| [1] | Prathap V, Suresh KUMAR, Nand Lal MEENA, Chirag MAHESHWARI, Monika DALAL, Aruna TYAGI. Phosphorus Starvation Tolerance in Rice Through a Combined Physiological, Biochemical and Proteome Analysis [J]. Rice Science, 2023, 30(6): 8-. |
| [2] | Serena REGGI, Elisabetta ONELLI, Alessandra MOSCATELLI, Nadia STROPPA, Matteo Dell’ANNO, Kiril PERFANOV, Luciana ROSSI. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Rice Engineered Lines [J]. Rice Science, 2023, 30(6): 6-. |
| [3] | Sundus ZAFAR, XU Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 4-. |
| [4] | Kankunlanach KHAMPUANG, Nanthana CHAIWONG, Atilla YAZICI, Baris DEMIRER, Ismail CAKMAK, Chanakan PROM-U-THAI. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 9-. |
| [5] | FAN Fengfeng, CAI Meng, LUO Xiong, LIU Manman, YUAN Huanran, CHENG Mingxing, Ayaz AHMAD, LI Nengwu, LI Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Rice Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 14-. |
| [6] | LIN Shaodan, YAO Yue, LI Jiayi, LI Xiaobin, MA Jie, WENG Haiyong, CHENG Zuxin, YE Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 10-. |
| [7] | Md. Forshed DEWAN, Md. AHIDUZZAMAN, Md. Nahidul ISLAM, Habibul Bari SHOZIB. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and South-East Asia: A Review [J]. Rice Science, 2023, 30(6): 5-. |
| [8] | Raja CHAKRABORTY, Pratap KALITA, Saikat SEN. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Chakhao Poireiton (A Pigmented Black Rice) in High-Fat High-Sugar induced Rats [J]. Rice Science, 2023, 30(6): 11-. |
| [9] | LI Qianlong, FENG Qi, WANG Heqin, KANG Yunhai, ZHANG Conghe, DU Ming, ZHANG Yunhu, WANG Hui, CHEN Jinjie, HAN Bin, FANG Yu, WANG Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 7-. |
| [10] | JI Dongling, XIAO Wenhui, SUN Zhiwei, LIU Lijun, GU Junfei, ZHANG Hao, Tom Matthew HARRISON, LIU Ke, WANG Zhiqin, WANG Weilu, YANG Jianchang. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 12-. |
| [11] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [12] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [13] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| [14] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| [15] | Zhang Guomei, Li Han, Liu Shanshan, Zhou Xuming, Lu Mingyang, Tang Liang, Sun Lihua. Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury [J]. Rice Science, 2023, 30(5): 473-485. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||