
Rice Science ›› 2016, Vol. 23 ›› Issue (6): 297-305.DOI: 10.1016/j.rsci.2016.07.002
• Orginal Article • Previous Articles Next Articles
Ashwini Narasimha, Sivarajan Sajeevan Radha, Udayakumar Makarla, Nalkur Nataraja Karaba
Received:2016-01-07
Accepted:2016-07-04
Online:2016-12-12
Published:2016-08-10
Ashwini Narasimha, Sivarajan Sajeevan Radha, Udayakumar Makarla, Nalkur Nataraja Karaba. Identification and Characterization of OsWRKY72 Variant in Indica Genotypes[J]. Rice Science, 2016, 23(6): 297-305.
Add to citation manager EndNote|Ris|BibTeX
| Name | Sequence (5′-3′) | Application |
| OsWRKY72-F1 | TCGATTGGTCGAGATGGAGAAC | Full length amplification |
| OsWRKY72-R1 | CAGTGTGGCATTGGCATTGA | Full length amplification |
| OsWRKY72a-F2 | CCCAAATGCACATCTACTCC | Expression of variant-a |
| OsWRKY72a-R2 | CAGTGTGGCATTGGCATTGA | Expression of variant-a |
| OsWRKY72b-F3 | CGAGAGGAGAACTTCCCGATACTCTTTGC | Expression of variant-b |
| OsWRKY72b-R3 | GTGAGGATGTGCTCGAAGTTGTCGTTGG | Expression of variant-b |
| OsLEA14-F | GCTACTCGCTGAAGAGCGCCGGG | Stress confirmation |
| OsLEA14-R | GATGGGGAGGTCGACGGTGAGGC | Stress confirmation |
| OsActin-F | TCCATAATGAAGTGTGATGT | Internal control |
| OsActin-R | GGACCTGACTCGTCATACTC | Internal control |
Table 1 Primers used in this study
| Name | Sequence (5′-3′) | Application |
| OsWRKY72-F1 | TCGATTGGTCGAGATGGAGAAC | Full length amplification |
| OsWRKY72-R1 | CAGTGTGGCATTGGCATTGA | Full length amplification |
| OsWRKY72a-F2 | CCCAAATGCACATCTACTCC | Expression of variant-a |
| OsWRKY72a-R2 | CAGTGTGGCATTGGCATTGA | Expression of variant-a |
| OsWRKY72b-F3 | CGAGAGGAGAACTTCCCGATACTCTTTGC | Expression of variant-b |
| OsWRKY72b-R3 | GTGAGGATGTGCTCGAAGTTGTCGTTGG | Expression of variant-b |
| OsLEA14-F | GCTACTCGCTGAAGAGCGCCGGG | Stress confirmation |
| OsLEA14-R | GATGGGGAGGTCGACGGTGAGGC | Stress confirmation |
| OsActin-F | TCCATAATGAAGTGTGATGT | Internal control |
| OsActin-R | GGACCTGACTCGTCATACTC | Internal control |
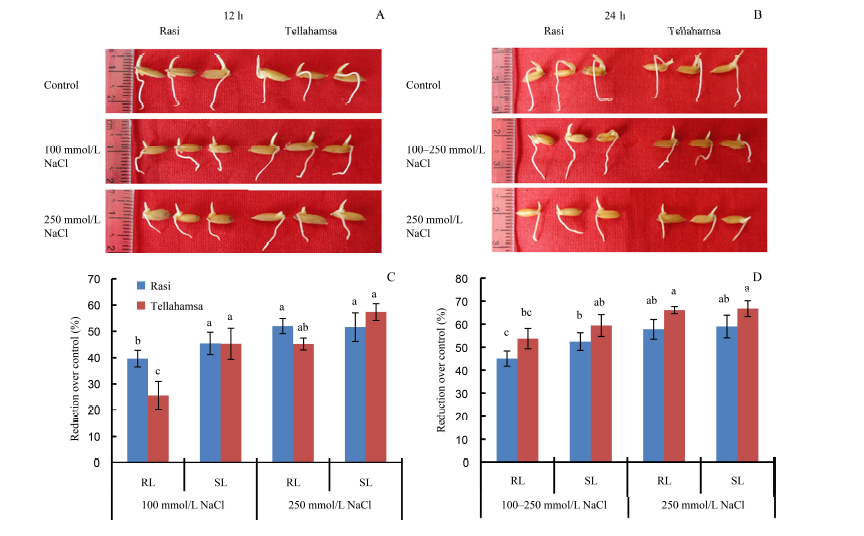
Fig. 1. Responses of Rasi and Tellahamsa seedlings to salinity stress. A, Phenotypes of Rasi and Tellahamsa seedlings at 12 h of NaCl stress; B, Phenotypes of Rasi and Tellahamsa seedlings at 24 h of NaCl stress; C, Percent reductions in root length (RL) and shoot length (SL) of Rasi and Tellahamsa over its control at 12 h; D, Percent reductions in root length (RL) and shoot length (SL) of Rasi and Tellahamsa over its control at 24 h. 100-250 mmol/L NaCl represents 100 mmol/L NaCl for 12 h followed by 250 mmol/L NaCl for 12 h. Significant differences at the 0.05 level are indicated by lowercase letters.
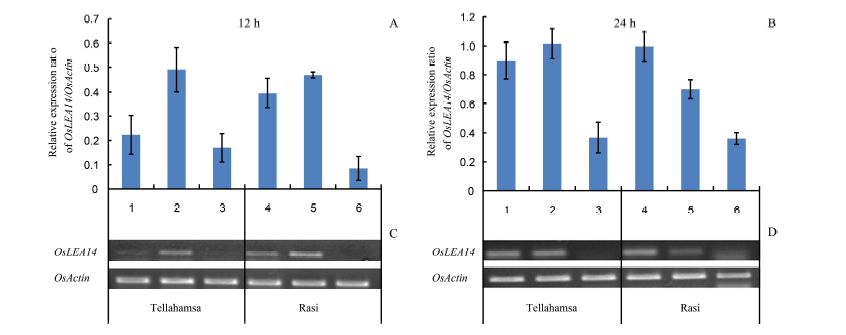
Fig. 2. Expression analyses of OsLEA14 in the seedlings of Rasi and Tellahamsa. Lanes 1, 2 and 3 correspond to expression in Tellahamsa, whereas lanes 4, 5 and 6 correspond to expression in Rasi. At 12 h, lanes 1 and 4 represent induction stress with 100 mmol/L NaCl; At 24 h, lanes 1 and 4 represent 100 mmol/L NaCl for 12 h followed by 250 mmol/L NaCl for 12 h; Lanes 2 and 5 are in lethal stress with 250 mmol/L NaCl, and lanes 3 and 6 are in control (distilled water) at 12 h and 24 h, respectively.
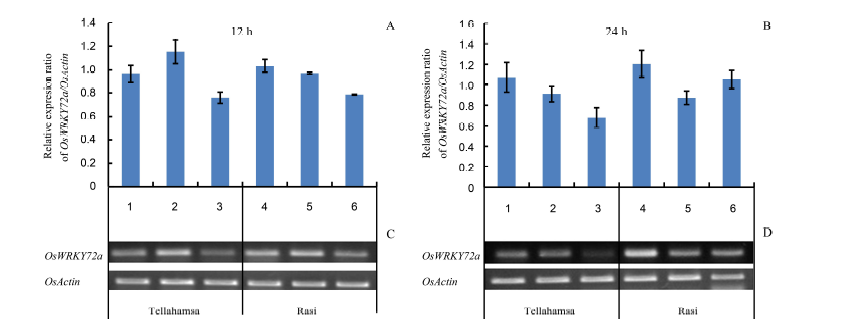
Fig. 3. Expression analyses of OsWRKY72a by semi-quantitative reverse transcriptase-PCR. Lanes 1, 2 and 3 correspond to expression in Tellahamsa, whereas lanes 4, 5 and 6 correspond to expression in Rasi. At 12 h, lanes 1 and 4 represent induction stress with 100 mmol/L NaCl; At 24 h, lanes 1 and 4 represent 100 mmol/L NaCl for 12 h followed by 250 mmol/L NaCl for 12 h; Lanes 2 and 5 are in lethal stress with 250 mmol/L NaCl, and lanes 3 and 6 are in control (distilled water) at 12 h and 24 h, respectively.
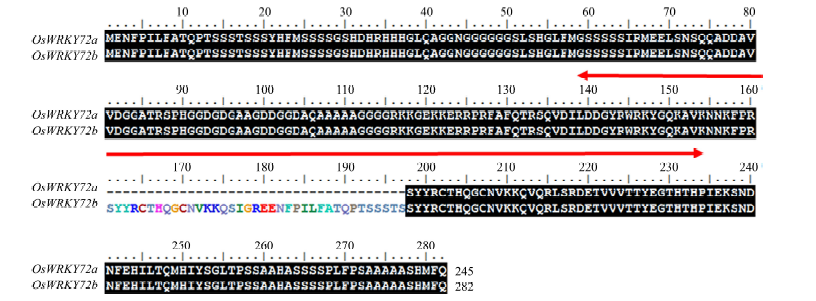
Fig. 4. Identification of OsWRKY72b in indica rice genotype. Alignments of OsWRKY72a and OsWRKY72b amino acid sequence. The region within red colored arrow indicates the WRKY domain, and additional sequence present in OsWRKY72b is represented by colored amino acids.
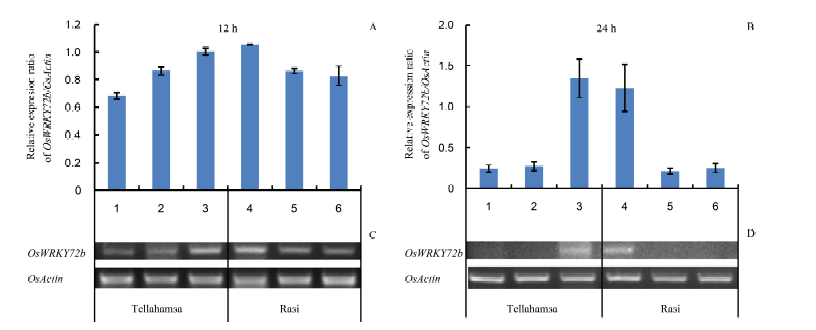
Fig. 5. Expression of OsWRKY72b in seedlings of Rasi and Tellahamsa under salinity stress. Lanes 1, 2 and 3 correspond to expression in Tellahamsa, whereas lanes 4, 5 and 6 correspond to expression in Rasi. At 12 h, lanes 1 and 4 represent induction stress with 100 mmol/L NaCl; At 24 h, lanes 1 and 4 represent 100 mmol/L NaCl for 12 h followed by 250 mmol/L NaCl for 12 h; Lanes 2 and 5 are in lethal stress with 250 mmol/L NaCl, and lanes 3 and 6 are in control (distilled water) at 12 h and 24 h, respectively.
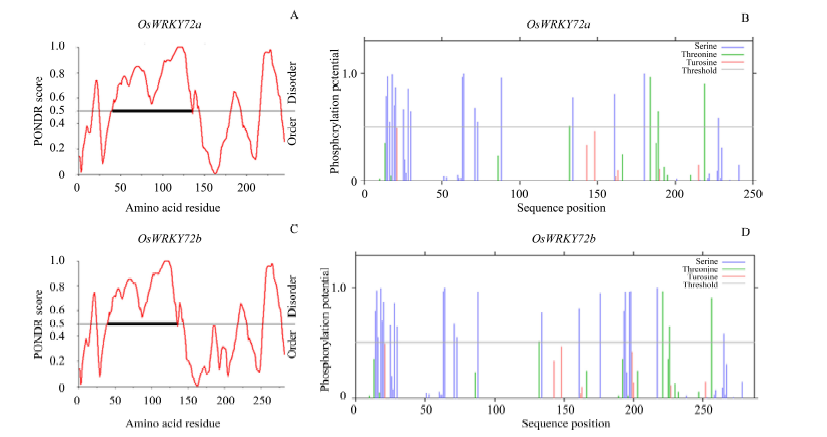
Fig. 6. In silico analysis of OsWKY72a and OsWKY72b. A and C, Predictor of natural disordered regions (PONDR) profiles for disorder; B and D, Phosphorylation site prediction using NetPhos 2 serv. The black rectangle on the threshold line indicates the region interacts with its binding partners.
| 1 | Agarwal P, Reddy M P, Chikara J.2011. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants.Mol Biol Rep, 38: 3883-3896. |
| 2 | Akua T, Berezin I, Shaul O.2010. The leader intron ofAtMHX can elicit, in the absence of splicing, low-level intron-mediated enhancement that depends on the internal intron sequence. BMC Plant Biol, 10: 93-102. |
| 3 | Akua T, Shaul O.2013. TheArabidopsis thaliana MHX gene includes an intronic element that boosts translation when localized in a 5′ UTR intron. J Exp Bot, 64: 4255-4270. |
| 4 | Blom N, Gammeltoft S, Brunak S.1999. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites.J Mol Biol, 294(5): 1351-1362. |
| 5 | Chen L G, Song Y, Li S J, Zhang L P, Zou C S, Yu D Q.2012. The role of WRKY transcription factors in plant abiotic stresses.Biochim Biophys Acta, 1819(2): 120-128. |
| 6 | Ciolkowski I, Wanke D, Birkenbihl R P, Somssich I E.2008. Studies on DNA-binding selectivity of WRKY transcription factors lend structural clues into WRKY-domain function.Plant Mol Biol, 68: 81-92. |
| 7 | Eulgem T, Rushton P J, Robatzek S, Somssich I E.2000. The WRKY superfamily of plant transcription factors.Trends Plant Sci, 5(5): 199-206. |
| 8 | Gianì S, Altana A, Campanoni P, Morello L, Breviario D.2009. In transgenic rice, alpha- and beta-tubulin regulatory sequences control GUS amount and distribution through intron mediated enhancement and intron dependent spatial expression.Transgenic Res, 18(2): 151-162. |
| 9 | Gomez K A, Gomez A A.1976. Statistical Procedures for Agricultural Research with Emphasis on Rice. Los Banos, the Philippines: International Rice Research Institute. |
| 10 | Hinderhofer K, Zentgraf U.2001. Identification of a transcription factor specifically expressed at the onset of leaf senescence.Planta, 213(3): 469-473. |
| 11 | Jayaprakash T L, Ramamohan G, Krishnaprasad B T, Ganeshkumar, Prasad T G, Mathew M K, Udayakumar M.1998. Genotypic variability in differential expression oflea2 and lea3 genes and proteins in response to salinity stress in fingermillet(Eleusine coracana Gaertn) and rice, 82: 513-522. |
| 12 | Johnson C S, Kolevski B, Smyth D R.2002. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene ofArabidopsis, encodes a WRKY transcription factor. Plant Cell, 14(6): 1359-1375. |
| 13 | Li R, Zhou J, Li D Y, Wang X M, Yang Y, Yu C L, Cheng Y, Yan C Q, Chen J P.2015. Expression ofOsWRKY7 in rice. Chin J Rice Sci, 29(6): 559-570. (in Chinese with English abstract) |
| 14 | Maeo K, Hayashi S, Kojima-Suzuki H, Morikami A, Nakamura K.2001. Role of conserved residues of the WRKY domain in the DNA-binding of tobacco WRKY family proteins.Biosci Biotech Bioch, 65(11): 2428-2436. |
| 15 | Pandey S P, Somssich I E.2009. The role of WRKY transcription factors in plant immunity.Plant Physiol, 150: 1648-1655. |
| 16 | Park S C, Kim Y H, Jeong J C, Kim C Y, Lee H S, Bang J W, Kwak S S.2011. Sweet potato late embryogenesis abundant 14 (IbLEA14) gene influences lignification and increases osmotic- and salt stress-tolerance of transgenic calli. Planta, 233(3): 621-634. |
| 17 | Popescu S C, Popescu G V, Bachan S, Zhang Z, Gerstein M, Snyder M, Dinesh-Kumar S P.2009. MAPK target networks inArabidopsis thaliana revealed using functional protein microarrays. Gene Dev, 23(1): 80-92. |
| 18 | Qiu Y P, Jing S J, Fu J, Li L, Yu D Q.2004. Cloning and analysis of expression profile of 13WRKY genes in rice. Chin Sci Bull, 49: 2159-2168. (in Chinese with English abstract) |
| 19 | Ramamoorthy R, Jiang S Y, Kumar N, Venkatesh P N, Ramachandran S.2008. A comprehensive transcriptional profiling of theWRKY gene family in rice under various abiotic and phytohormone treatments. Plant Cell Physiol, 49(6): 865-879. |
| 20 | Romero P, Obradovic Z, Dunker A K.1997. Sequence data analysis for long disordered regions prediction in the calcineurin family.Genome Inform Workshop Genome Inform, 8: 110-124. |
| 21 | Sajeevan R S, Shivanna M B, Nataraja K N.2014. An efficient protocol for total RNA isolation from healthy and stressed tissues of mulberry (Morus sp.) and other species. Am J Plant Sci, 5(13): 2057-2065. |
| 22 | Sambrook J, Fritsch E F, Maniatis T.1989. Molecular Cloning: A Laboratory Manual. 2nd ed. New York, USA: Cold Spring Harbor Laboratory Press. |
| 23 | Shen H S, Liu C T, Zhang Y, Meng X P, Zhou X, Chu C C, Wang X P.2012. OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice.Plant Mol Biol, 80(3): 241-253. |
| 24 | Singh S, Cornilescu C C, Tyler R C, Cornilescu G, Tonelli M, Lee M S, Markley J L.2005. Solution structure of a late embryogenesis abundant protein (LEA14) fromArabidopsis thaliana, a cellular stress-related protein. Protein Sci, 14(10): 2601-2609. |
| 25 | Song Y, Ai C R, Jing S J, Yu D Q.2010a. Research progress on function analysis of riceWRKY gene. Rice Sci, 17(1): 60-72. |
| 26 | Song Y, Chen L G, Zhang L P, Yu D Q.2010b. Overexpression ofOsWRKY72 gene interferes in the abscisic acid signal and auxin transport pathway of Arabidopsis. J Biosciences, 35(3): 459-471. |
| 27 | Song Y, Jing S J, Yu D Q.2009. Overexpression of the stress inducedOsWRKY08 improves the osmotic stress tolerance in Arabidopsis. Chin Sci Bull, 54: 4671-4678. (in Chinese with English abstract) |
| 28 | Uma S, Ravishankar K V, Prasad T G, Reid J L, Udaya Kumar M.1993. Abscisic acid-responsive proteins induce salinity stress tolerance in finger millet (Eleusine coracana Gaertn.) seedlings. Curr Sci, 65: 549-554. |
| 29 | Wu K L, Guo Z J, Wang H H, Li J.2005. The WRKY family of transcription factors in rice andArabidopsis and their origins. DNA Res, 12(1): 9-26. |
| 30 | Wu X L, Shiroto Y, Kishitani S, Ito Y, Toriyama K.2009. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressingOsWRKY11 under the control of HSP101 promoter. Plant Cell Rep, 28(1): 21-30. |
| 31 | Xie Z, Zhang Z L, Zou X L, Huang J, Ruas P, Thompson D, Shen Q J.2005. Annotations and functional analyses of the riceWRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol, 137: 176-189. |
| 32 | Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S.2008. Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains.Plant Physiol Bioch, 46(3): 394-401. |
| 33 | (Managing Editor: Wang Caihong) |
| [1] | B. M. Lokeshkumar, S. L. Krishnamurthy, Suman Rathor, Arvinder Singh Warriach, N. M. Vinaykumar, B. M. Dushyanthakumar, Parbodh Chander Sharma. Morphophysiological Diversity and Haplotype Analysis of Saltol QTL Region in Diverse Rice Landraces for Salinity Tolerance [J]. Rice Science, 2023, 30(4): 306-320. |
| [2] | M. Iqbal R. Khan, Sarika Kumari, Faroza Nazir, Risheek Rahul Khanna, Ravi Gupta, Himanshu Chhillar. Defensive Role of Plant Hormones in Advancing Abiotic Stress-Resistant Rice Plants [J]. Rice Science, 2023, 30(1): 15-35. |
| [3] | Shalini Pulipati, Suji Somasundaram, Nitika Rana, Kavitha Kumaresan, Mohamed Shafi, Peter Civáň, Gothandapani Sellamuthu, Deepa Jaganathan, Prasanna Venkatesan Ramaravi, S. Punitha, Kalaimani Raju, Shrikant S. Mantri, R. Sowdhamini, Ajay Parida, Gayatri Venkataraman. Diversity of Sodium Transporter HKT1;5 in Genus Oryza [J]. Rice Science, 2022, 29(1): 31-46. |
| [4] | Tianqiao Song, Xiong Zhang, You Zhang, Dong Liang, Jiaoling Yan, Junjie Yu, Mina Yu, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Yongfeng Liu. Genome-Wide Identification of Zn2Cys6 Class Fungal-Specific Transcription Factors (ZnFTFs) and Functional Analysis of UvZnFTF1 in Ustilaginoidea virens [J]. Rice Science, 2021, 28(6): 567-578. |
| [5] | Chuxin Wang, Chengchao Zhu, Yu Zhou, Min Xiong, Jindong Wang, Huang Bai, Chenya Lu, Changquan Zhang, Qiaoquan Liu, Qianfeng Li. OsbZIP09, a Unique OsbZIP Transcription Factor of Rice, Promotes Rather Than Suppresses Seed Germination by Attenuating Abscisic Acid Pathway [J]. Rice Science, 2021, 28(4): 358-367. |
| [6] | Fuad Anshori Muhammad, Sapta Purwoko Bambang, Saraswati Dewi Iswari, Wahyuning Ardie Sintho, Bayuardi Suwarno Willy. A New Approach to Select Doubled Haploid Rice Lines under Salinity Stress Using Indirect Selection Index [J]. Rice Science, 2021, 28(4): 368-378. |
| [7] | Baoxiang Wang, Yan Liu, Yifeng Wang, Jingfang Li, Zhiguang Sun, Ming Chi, Yungao Xing, Bo Xu, Bo Yang, Jian Li, Jinbo Liu, Tingmu Chen, Zhaowei Fang, Baiguan Lu, Dayong Xu, Kazeem Bello Babatunde. OsbZIP72 Is Involved in Transcriptional Gene-Regulation Pathway of Abscisic Acid Signal Transduction by Activating Rice High-Affinity Potassium Transporter OsHKT1;1 [J]. Rice Science, 2021, 28(3): 257-267. |
| [8] | Bhatt Tarun, Sharma Aditi, Puri Sanjeev, Priya Minhas Anu. Salt Tolerance Mechanisms and Approaches: Future Scope of Halotolerant Genes and Rice Landraces [J]. Rice Science, 2020, 27(5): 368-383. |
| [9] | Yongqi He, Jia Zhao, Defeng Feng, Zhibo Huang, Jiaming Liang, Yufei Zheng, Jinping Cheng, Jifeng Ying, Zhoufei Wang. RNA-Seq Study Reveals AP2-Domain-Containing Signalling Regulators Involved in Initial Imbibition of Seed Germination in Rice [J]. Rice Science, 2020, 27(4): 302-314. |
| [10] | Xixu Peng, Ting Xiao, Jiao Meng, Zong Tao, Dinggang Zhou, Xinke Tang, Haihua Wang. Differential Expression of Rice Valine-Qlutamine Gene Family in Response to Nitric Oxide and Regulatory Circuit of OsVQ7 and OsWRKY24 [J]. Rice Science, 2020, 27(1): 10-20. |
| [11] | Elisa B. Gerona Maria, P. Deocampo Marjorie, A. Egdane James, M. Ismail Abdelbagi, L. Dionisio-Sese Maribel. Physiological Responses of Contrasting Rice Genotypes to Salt Stress at Reproductive Stage [J]. Rice Science, 2019, 26(4): 207-219. |
| [12] | F. Aala Jr Wilson, B. Gregorio Glenn. Morphological and Molecular Characterization of Novel Salt-Tolerant Rice Germplasms from the Philippines and Bangladesh [J]. Rice Science, 2019, 26(3): 178-188. |
| [13] | Bo Wang, Zhaohui Zhong, Huanhuan Zhang, Xia Wang, Binglin Liu, Lijia Yang, Xiangyan Han, Deshui Yu, Xuelian Zheng, Chunguo Wang, Wenqin Song, Chengbin Chen, Yong Zhang. Targeted Mutagenesis of NAC Transcription Factor Gene, OsNAC041, Leading to Salt Sensitivity in Rice [J]. Rice Science, 2019, 26(2): 98-108. |
| [14] | Naga Bheema Lingeswara Reddy Inja, Kim Sung-Mi, Kim Beom-Ki, Yoon In-Sun, Kwon Taek-Ryoun. Identification of Rice Accessions Associated with K+/Na+ Ratio and Salt Tolerance Based on Physiological and Molecular Responses [J]. Rice Science, 2017, 24(6): 360-364. |
| [15] | Naga Bheema Lingeswara Reddy Inja, Kim Beom-Ki, Yoon In-Sun, Kim Kyung-Hwan, Kwon Taek-Ryoun. Salt Tolerance in Rice: Focus on Mechanisms and Approaches [J]. Rice Science, 2017, 24(3): 123-144. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||